Pending Review
Microbiomes contribute to multiple ecosystem services by transforming organic matter in soil. Extreme shifts in the environment, such as drying-rewetting cycles during drought, can impact microbial metabolism of organic matter by altering their physiology and function. These physiological responses are regulated in part by lipids that are responsible for regulating interactions between cells and the environment. Despite this critical role in regulating microbial response to stress, little is known about microbial lipids and metabolites in soil or how they influence phenotypes that are expressed under drying- rewetting cycles. To address this knowledge gap, we conducted a soil incubation experiment to simulate soil drying during a summer drought of an arid grassland, then measured the response of the soil lipidome and metabolome during the first 3 h after wet-up. See resulting data package DOI: 10.25584/WAOmicsLA1/1773648.
IMPACT
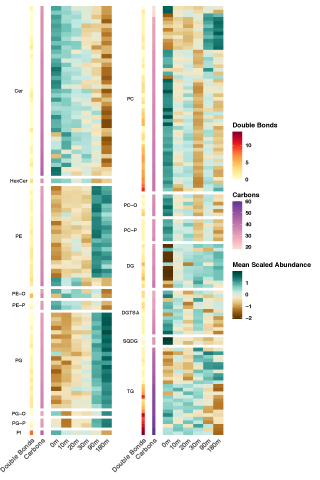
This study revealed specific changes in lipids and metabolites that are indicative of stress adaptation, substrate use, and cellular recovery during soil drying and subsequent rewetting. Drought induced nutrient limitation was reflected in the lipidome and polar metabolome, both of which rapidly shifted (within hours) upon rewet. Reduced nutrient access in dry soil caused the replacement of glycerophospholipids with phosphorus-free lipids and impeded resource-expensive osmolyte accumulation. Elevated levels of ceramides and lipids with long chain polyunsaturated fatty acids, in dry soil suggests that lipids play an important role in fungal drought tolerance. Increasing abundance of bacterial glycerophospholipids and triacylglycerols with fatty acids typical of bacteria and polar metabolites suggest metabolic recovery in representative bacteria once the environmental conditions are conducive for growth. These results underscore the importance of the soil lipidome as a robust indicator of microbial community responses, especially at the short time scales of cell-environment reactions.
AFFILIATED INSTITUTIONS
- Earth and Biological Sciences Directorate, Pacific Northwest National Laboratory, Richland, WA, USA (https://ror.org/05h992307)
- National Security Directorate, Pacific Northwest National Laboratory, Richland, WA, USA (https://ror.org/05h992307)
- Department of Molecular Microbiology and Immunology, Oregon Health & Science University, Portland, OR, USA (https://ror.org/009avj582)
- Department of Agronomy, Iowa State University, Ames, IA, USA (https://ror.org/04rswrd78)
Projects (1)
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
45