Filter results
Content type
Tags
- (-) Metagenomics (9)
- (-) Type 1 Diabetes (6)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (30)
- Multi-Omics (28)
- Viruses (26)
- Health (23)
- Omics (23)
- Virus (23)
- Soil Microbiology (21)
- MERS-CoV (18)
- Mus musculus (18)
- sequencing (13)
- West Nile virus (13)
- Ebola (11)
- Genomics (11)
- Influenza A (11)
- Mass Spectrometry (8)
- Microbiome (8)
- Resource Metadata (8)
- High Throughput Sequencing (7)
- Microarray (7)
- Fungi (6)
- Human Interferon (6)
- Imaging (6)
"DNA Viral Diversity, Abundance, and Functional Potential Vary across Grassland Soils with a Range of Historical Moisture Regimes" Soil viruses are abundant, but the influence of the environment and climate on soil viruses remains poorly understood. Here, we addressed this gap by comparing the...
Category
The Human Islet Research Network (HIRN) is a large consortia with many research projects focused on understanding how beta cells are lost in type 1 diabetics (T1D) with a goal of finding how to protect against or replace the loss of functional beta cells. The consortia has multiple branches of...
Datasets
0
The Environmental Determinants of Diabetes in the Young (TEDDY) study is searching for factors influencing the development of type 1 diabetes (T1D) in children. Research has shown that there are certain genes that correlate to higher risk of developing T1D, but not all children with these genes...
Datasets
1
The Diabetes Autoimmunity Study in the Young (DAISY) seeks to find environmental factors that can trigger the development of type 1 diabetes (T1D) in children. DAISY follows children with high-risk of developing T1D based on family history or genetic markers. Genes, diets, infections, and...
Datasets
1
Machine learning is a core technology that is rapidly advancing within type 1 diabetes (T1D) research. Our Human Islet Research Network (HIRN) grant is studying early cellular response initiating β cell stress in T1D through the generation of heterogenous low- and high-throughput molecular...
Datasets
3
Category
Category
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. WA-TmG.2.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG2/1770324 Soil samples were collected in triplicate in the fall of 2017 across the three grassland...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. KS-TmG.2.0 (Metagenome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KSTmG2/1770332 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. IA-TmG.2.0 (Metagenome, IA). [Data Set] PNNL DataHub. https://doi.org/10.25584/IATmG2/1770333 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
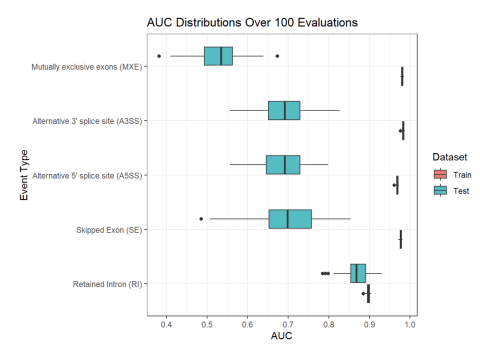
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
A total of 172 children from the DAISY study with multiple plasma samples collected over time, with up to 23 years of follow-up, were characterized via proteomics analysis. Of the children there were 40 controls and 132 cases. All 132 cases had measurements across time relative to IA. Sampling was...