The rhizosphere represents a dynamic and complex interface between plant hosts and the microbial community found in the surrounding soil. While it is recognized that manipulating the rhizosphere has the potential to improve plant fitness and health, engineering the rhizosphere microbiome through...
Filter results
Category
- (-) Earth System Science (136)
- (-) Microbiome Science (42)
- (-) Computational Research (24)
- (-) Renewable Energy (5)
- Scientific Discovery (308)
- Biology (199)
- Human Health (103)
- Integrative Omics (74)
- National Security (21)
- Computing & Analytics (14)
- Chemistry (10)
- Energy Resiliency (10)
- Data Analytics & Machine Learning (9)
- Materials Science (7)
- Computational Mathematics & Statistics (6)
- Visual Analytics (6)
- Chemical & Biological Signatures Science (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Coastal Science (4)
- Ecosystem Science (4)
- Data Analytics & Machine Learning (3)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Omics (23)
- Soil Microbiology (21)
- sequencing (13)
- Genomics (12)
- High Throughput Sequencing (9)
- Metagenomics (9)
- PerCon SFA (9)
- Microbiome (8)
- Mass Spectrometry (7)
- Fungi (6)
- Imaging (6)
- Machine Learning (6)
- Type 1 Diabetes (6)
- Autoimmunity (5)
- Mass Spectrometer (5)
- Sequencer System (5)
- Synthetic Biology (5)
- Biomarkers (4)
- metagenomics (4)
- Microscopy (4)
- Molecular Profiling (4)
- Proteomics (4)
- RNA Sequence Analysis (4)
- Sequencing (4)
- Software Data Analysis (4)
- soil microbiology (4)
- Spectroscopy (4)
- Climate Change (3)
- IAREC (3)
- Viruses (3)
Agriculture is the largest source of greenhouse gases (GHG) production. Conversion of nitrogen fertilizers into more reduced forms by microbes through a process known as biological nitrification drives GHG production, enhances proliferation of toxic algal blooms, and increases cost of crop...
Metabolite exchange between plant roots and their associated rhizosphere microbiomes underpins plant growth promotion by microbes. Sorghum bicolor is a cereal crop that feeds animals and humans and is used for bioethanol production. Its root tips exude large amounts of a lipophilic benzoquinone...
A major challenge in biotechnology and biomanufacturing is the identification of a set of biomarkers for perturbations and metabolites of interest. Here, we develop a data-driven, transcriptome-wide approach to rank perturbation-inducible genes from time-series RNA sequencing data for the discovery...
The Sequel II System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by PacBio , and is designed for high throughput, production-scale sequencing laboratories. Originally released in 2015, the Sequel system provides Single Molecule, Real-Time (SMRT) sequencing core...
The Human Islet Research Network (HIRN) is a large consortia with many research projects focused on understanding how beta cells are lost in type 1 diabetics (T1D) with a goal of finding how to protect against or replace the loss of functional beta cells. The consortia has multiple branches of...
Datasets
0
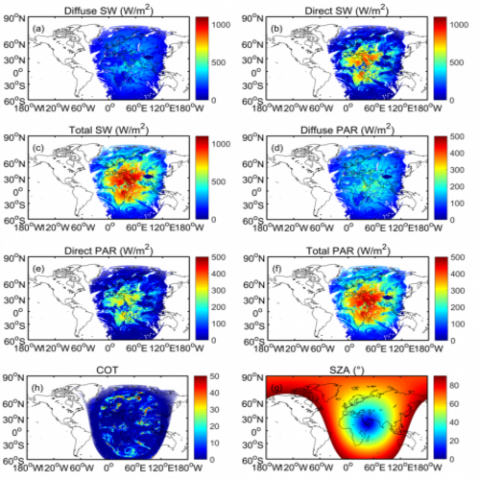
This project is an interdisciplinary collaboration supported by US DOE Office of Science's Scientific Discovery through Advanced Computing (SciDAC) program. The project addresses a crucial but largely overlooked source of error in the Energy Exascale Earth System Model (E3SM) and other atmosphere...
Category
Datasets
2
Category
Datasets
1
Category
Datasets
1
Category
Datasets
1
Category
Datasets
54
The influence of tidal inundation dynamics on below ground carbon pools is poorly understood across coastal terrestrial-aquatic interface (TAI) ecosystems. The dynamic environmental conditions of tidally-influenced landscapes, the chemically complex nature of carbon compounds, the diverse nature of...
Category
Datasets
3
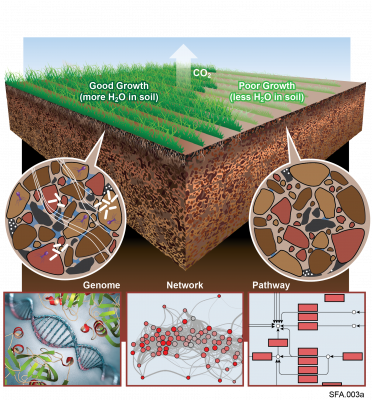
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
23
pmartR Software Overview The pmartR package provides a single software tool for QC (filtering and normalization), exploratory data analysis (EDA), and statistical analysis (robust to missing data) and includes numerous visualization capabilities of mass spectrometry (MS) omics data (proteomic...
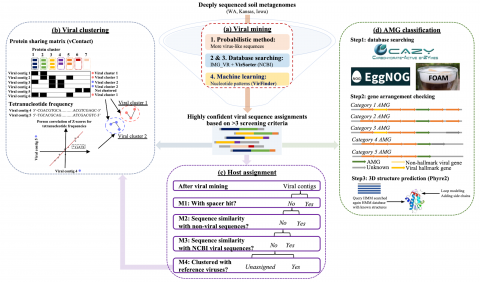
The following R source code was used for plotting figures of the viral communities detected from three grasslands soil metagenomes with a historical precipitation gradient ( WA-TmG.2.0 , KS-TmG.2.0 , IA-TmG.2.0 ) from project publication 'DNA viral diversity, abundance and functional potential vary...