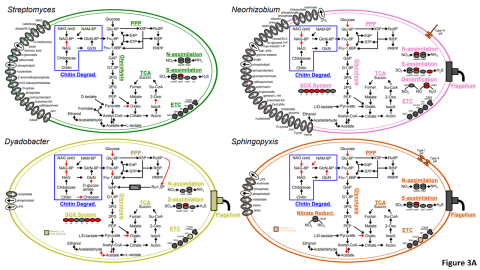
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Filter results
Content type
Tags
- (-) Metagenomics (10)
- (-) High Throughput Sequencing (6)
- (-) Molecular Profiling (4)
- (-) Sequencer System (4)
- Omics (22)
- Soil Microbiology (22)
- sequencing (13)
- Genomics (10)
- Microbiome (8)
- Fungi (6)
- Imaging (6)
- Mass Spectrometry (6)
- Type 1 Diabetes (6)
- Autoimmunity (5)
- Machine Learning (5)
- Mass Spectrometer (5)
- Biomarkers (4)
- metagenomics (4)
- Microscopy (4)
- Sequencing (4)
- soil microbiology (4)
- Spectroscopy (4)
- Viruses (4)
- Climate Change (3)
- IAREC (3)
- Mass spectrometry-based Omics (3)
- metabolomics (3)
- PerCon SFA (3)
- Proteomics (3)
- Synthetic Biology (3)
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. KS-TmG.2.0 (Metagenome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KSTmG2/1770332 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. WA-TmG.2.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG2/1770324 Soil samples were collected in triplicate in the fall of 2017 across the three grassland...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. IA-TmG.2.0 (Metagenome, IA). [Data Set] PNNL DataHub. https://doi.org/10.25584/IATmG2/1770333 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Category
Category
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
"Moisture modulates soil reservoirs of active DNA and RNA viruses" Soil is known to harbor viruses, but the majority are uncharacterized and their responses to environmental changes are unknown. Here, we used a multi-omics approach (metagenomics, metatranscriptomics and metaproteomics) to detect...
The Diabetes Autoimmunity Study in the Young (DAISY) seeks to find environmental factors that can trigger the development of type 1 diabetes (T1D) in children. DAISY follows children with high-risk of developing T1D based on family history or genetic markers. Genes, diets, infections, and...
Datasets
1
The Human Islet Research Network (HIRN) is a large consortia with many research projects focused on understanding how beta cells are lost in type 1 diabetics (T1D) with a goal of finding how to protect against or replace the loss of functional beta cells. The consortia has multiple branches of...
Datasets
0
The Environmental Determinants of Diabetes in the Young (TEDDY) study is searching for factors influencing the development of type 1 diabetes (T1D) in children. Research has shown that there are certain genes that correlate to higher risk of developing T1D, but not all children with these genes...
Datasets
1
Machine learning is a core technology that is rapidly advancing within type 1 diabetes (T1D) research. Our Human Islet Research Network (HIRN) grant is studying early cellular response initiating β cell stress in T1D through the generation of heterogenous low- and high-throughput molecular...
Datasets
3
The Sequel II System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by PacBio , and is designed for high throughput, production-scale sequencing laboratories. Originally released in 2015, the Sequel system provides Single Molecule, Real-Time (SMRT) sequencing core...
The Illumina HiSeq 4000 System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for high throughput, production-scale sequencing laboratories. Built off the HiSeq 2500 System and harnessing the patterned flow cell technology originally...