Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from KPBS field site in Manhattan, KS. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Filter results
Category
- (-) Biology (9)
- Scientific Discovery (9)
- Earth System Science (4)
- Microbiome Science (3)
- Computational Research (2)
- Data Analytics & Machine Learning (2)
- Human Health (2)
- Computational Mathematics & Statistics (1)
- Computational Mathematics & Statistics (1)
- Computing & Analytics (1)
- Data Analytics & Machine Learning (1)
- Integrative Omics (1)
- National Security (1)
Content type
Tags
- (-) Sequencing (3)
- Virology (63)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (25)
- Multi-Omics (23)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (14)
- Health (13)
- Soil Microbiology (13)
- Virus (13)
- West Nile virus (11)
- Ebola (9)
- Influenza A (9)
- sequencing (9)
- Resource Metadata (8)
- Metagenomics (7)
- Microarray (5)
- Genomics (4)
- Human Interferon (4)
- Microbiome (4)
- Omics (4)
- Fungi (3)
- IAREC (3)
- metagenomics (3)
- soil microbiology (3)
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from COBS field site in Boone County, IA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from IAREC field site in Prosser, WA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
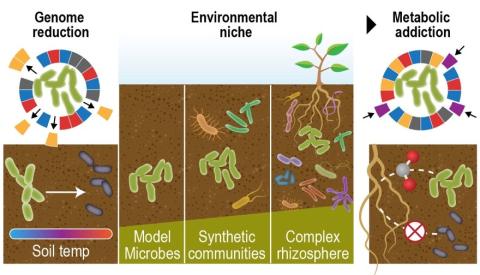
Last updated on 2023-05-02T18:08:23+00:00 by LN Anderson Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68" Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for...
Category
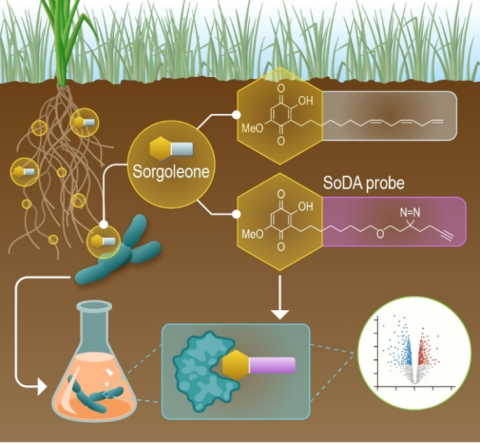
Last updated on 2024-04-19T19:12:08+00:00 by LN Anderson PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...
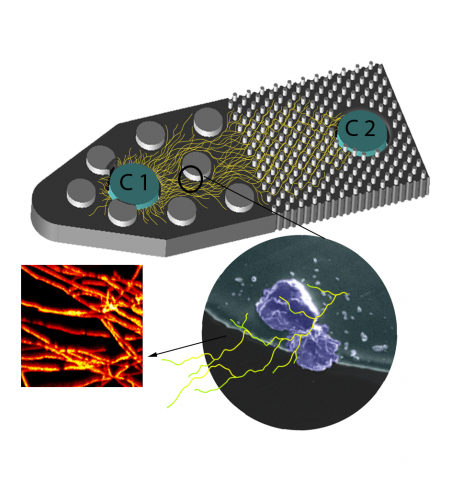
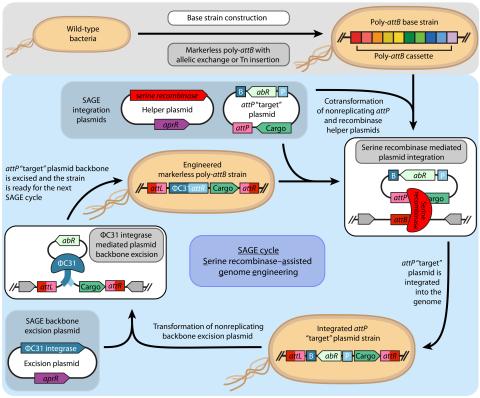
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
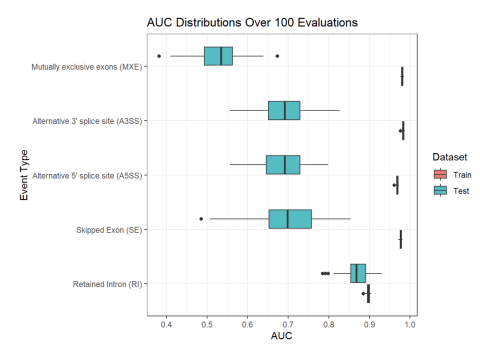
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...