These GCAM v4.3 SSP-RCP-GCM Output Databases are made available under the Open Data Commons Attribution License: http://opendatacommons.org/licenses/by/1.0/ . GCAM v4.3 SSP-RCP-GCM plausible solution databases. Supplemental dataset to: Graham N.T., M.I. Hejazi, M. Chen, E. Davies, J.A. Edmonds, S.H...
Filter results
Category
- Scientific Discovery (85)
- Biology (83)
- Human Health (66)
- Integrative Omics (50)
- Earth System Science (15)
- Computational Research (4)
- Data Analytics & Machine Learning (3)
- Microbiome Science (3)
- Computational Mathematics & Statistics (2)
- National Security (2)
- Chemical & Biological Signatures Science (1)
- Computing & Analytics (1)
- Data Analytics & Machine Learning (1)
- High-Performance Computing (1)
- Weapons of Mass Effect (1)
Content type
Dataset Type
- (-) Omics (86)
- (-) Computed Analysis (3)
- Transcriptomics (51)
- Proteomics (39)
- Sequencing (39)
- Metabolomics (28)
- Lipidomics (26)
- Simulation Results (17)
- Amplicon (16S, ITS) (15)
- Metagenomics (9)
- Microarray (9)
- Genomics (4)
- ChIP-seq (2)
- Geospacial (2)
- Staff (2)
- Cybersecurity (1)
- Electromagnetic Spectrum (1)
- Mass Spectra (1)
- Microscopy (1)
- Phenomics (1)
- Whole genome sequencing (WGS) (1)
Tags
- Virology (61)
- Immune Response (46)
- Time Sampled Measurement Datasets (43)
- Differential Expression Analysis (41)
- Gene expression profile data (40)
- Homo sapiens (28)
- Mass spectrometry data (25)
- Multi-Omics (23)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (14)
- Health (13)
- Virus (13)
- West Nile virus (11)
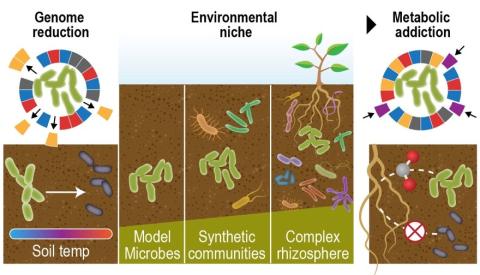
- Soil Microbiology (10)
- Ebola (9)
- Influenza A (7)
- Metagenomics (7)
- sequencing (7)
- Resource Metadata (6)
- Microarray (5)
- Human Interferon (4)
- Omics (4)
- Genomics (3)
- metagenomics (3)
- Microbiome (3)
- PerCon SFA (3)
- Response to type I interferon (3)
- Sequencing (3)
- soil microbiology (3)
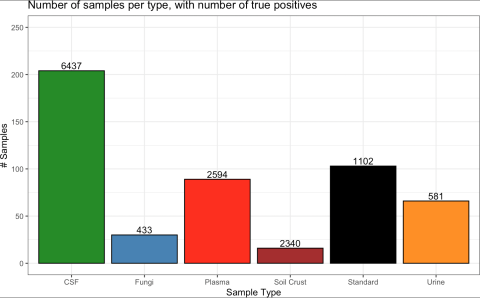
Data comprised of 204, 103, 89, 66, 30, and 16 gas chromatography mass spectrometry (GC-MS) runs from human cerebrospinal fluid (CSF), standards, human blood plasma, human urine, fungi species (A. niger, A. nidulans, T. reesei), and soil crust, respectively Across 4,523,251 candidate matches, 13,487...
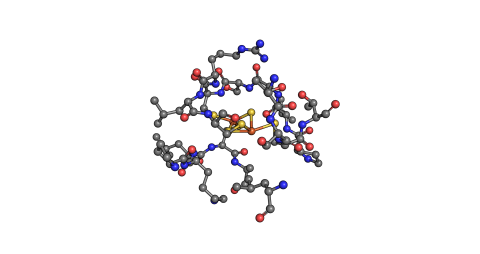
A pre-trained convolutional neural network was fine-tuned for three separate classification tasks, distinguishing 2D images of: 1) single amino acids, 2) protein structural ball and stick images of metalloproteins, and 3) protein structural ball and stick images of metalloenzymes with the metal...
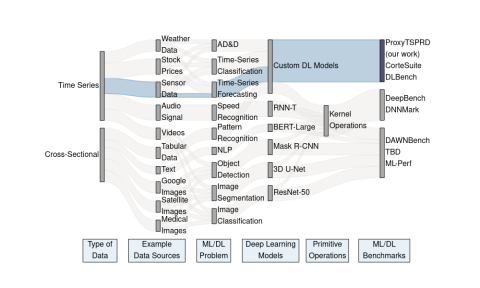
ProxyTSPRD profiles are collected using NVIDIA Nsight Systems version 2020.3.2.6-87e152c and capture computational patterns from training deep learning-based time-series proxy-applications on four different levels: models (Long short-term Memory and Convolutional Neural Network), DL frameworks...
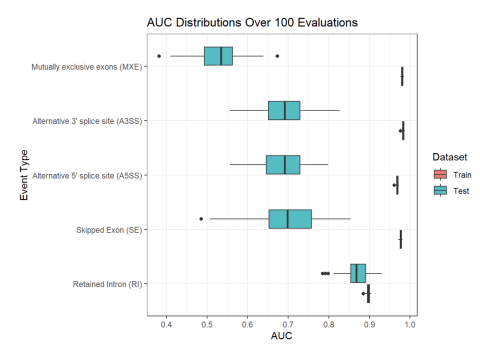
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
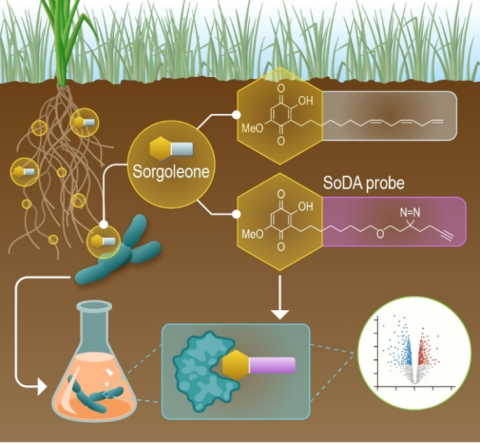
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
Last updated on 2024-04-19T19:12:08+00:00 by LN Anderson PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
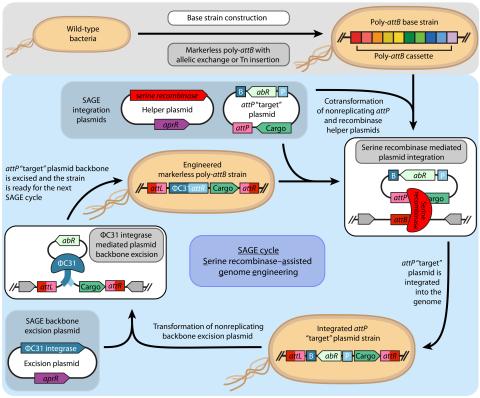
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
A total of 172 children from the DAISY study with multiple plasma samples collected over time, with up to 23 years of follow-up, were characterized via proteomics analysis. Of the children there were 40 controls and 132 cases. All 132 cases had measurements across time relative to IA. Sampling was...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
To understand how chemical exposure can impact health, researchers need tools that capture the complexities of personal chemical exposure. In practice, fine particulate matter (PM2.5) air quality index (AQI) data from outdoor stationary monitors and Hazard Mapping System (HMS) smoke density data...
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...