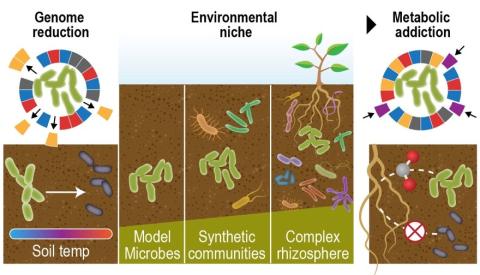
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson PerCon SFA Project Publication Experimental Data Catalog The Persistence Control of Engineered Functions in Complex Soil Microbiomes Project (PerCon SFA) at Pacific Northwest National Laboratory ( PNNL ) is a Genomic Sciences Program...
Datasets
3