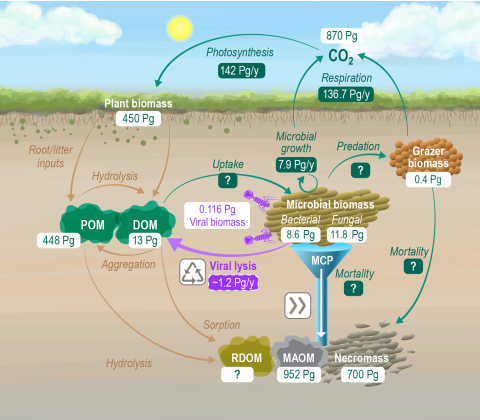
Original data published and available from Zenodo at: https://doi.org/10.5281/zenodo.13839485 Please cite as : Zimmerman, A., & Hofmockel, K. (2024). Global terrestrial carbon pools and fluxes [Data set]. In Global Change Biology. Zenodo. https://doi.org/10.5281/zenodo.13839485 This file contains a...
Filter results
Category
- (-) Scientific Discovery (404)
- (-) Chemical & Biological Signatures Science (12)
- Biology (287)
- Earth System Science (167)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (26)
- Mass Spectrometry (25)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
- Post-Translational Modifications (8)
- Microarray (7)
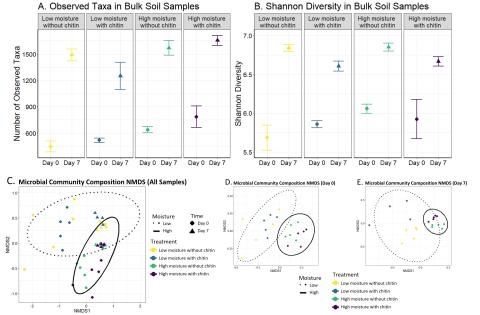
Please cite as : Rodríguez-Ramos, Josué, Natalie Sadler, Elias K. Zegeye, Yuliya Farris, Samuel Purvine, Sneha Couvillion, William C. Nelson, and Kirsten Hofmockel. Environmental matrix and moisture are key determinants of microbial phenotypes expressed in a reduced complexity soil-analog. 2024....
Please cite as : Nicholas J Reichart, Sheryl Bell, Vanessa A. Garayburu-Caruso, Natalie Sadler, Sharon Zhao, Kirsten Hofmockel. Chitin Decomposition 16S rRNA gene fastq. 2024 . [Data Set] PNNL DataHub. 10.25584/2475041. This data is published under a CC0 license . The authors encourage data reuse...
Category
Currently pending public release.
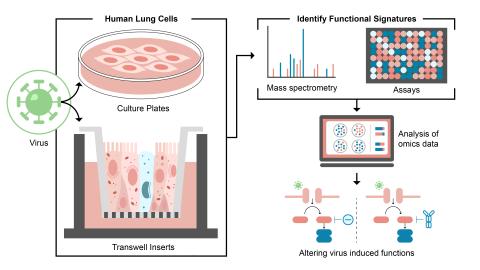
Dataset Description The purpose of this experiment was to evaluate the human host cellular response to wild type human coronavirus strain 229E (HCoV-229E) infection. Sample data was obtained for mock-infected and HCoV-229E infected immortalized human lung epithelial cells (A549) (MOI 5) and...
Created on 2024-10-16T17:27:40+00:00 by LN Anderson ; Last updated 2025-08-01T15:13:46+00:00. S. elongatus PCC 7942 LiP and TPP Structural Proteomics (JM-PB-DP3) The purpose of this experiment was to investigate structural alterations in proteins involved in central carbon metabolism and...
Currently pending public release.
Dataset Description The purpose of this experiment was to evaluate how wild-type Human coronavirus strain 229E (HCoV-299E) infection alters chromatin accessibility in infected cells. Sample data was obtained from mock-infected cells, UV-inactivated virus treated cells, and replication competent HCoV...
Category
Currently pending public release.
Category
Dataset Description The purpose of this experiment was to evaluate the human host cellular response to wild-type Human coronavirus strain 229E (HCoV-299E) infection. Sample data was obtained for mock and infected (MOI 3) primary human airway epithelial cells with and without macrophages and grown in...
Category
Data Description Co-cultured S. elongatus PCC 7942 CscB/SPS and R. toruloides IFO0880 presented as an effective photosynthesis-driven biofuel production platform. The goal of this experiment was to understand the molecular mechanism of this co-culture system at redox post-translational modification...
Category
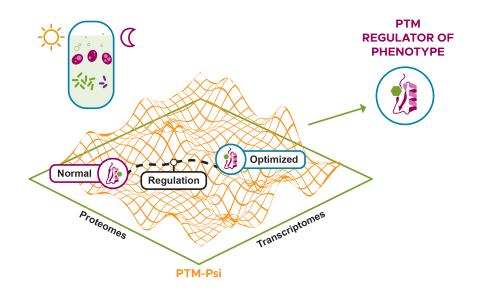
Dataset Description The purpose of this experiment was to examine the redox proteome of S. elongatus PCC 7942 CscB/SPS under different light conditions (light and dark) and culture conditions (dense and dilute optical density). Samples were processed using a resin-assisted capture (RAC) workflow...
Category
Dataset Description The purpose of this experiment was to evaluate the human host cellular response to wild-type Human coronavirus strain 229E (HCoV-229E) infection. Sample data was obtained for mock and infected immortalized human lung epithelial cells (A549) (MOI 5) nuclear extracts, immortalized...
Dataset Description The purpose of this experiment was to evaluate the human host cellular response to wild-type Human coronavirus strain 229E (HCoV-229E) infection. Sample data was obtained for mock and infected immortalized human lung epithelial cells (A549) (MOI 5), immortalized human lung...
Category
Last updated on 2025-01-31T00:37:40+00:00 by LN Anderson High-density Lipoprotein (HDL) Structure and Function (JM-DP1) The purpose of this experiment was to investigate how the interactions between APOA1 and APOA2 on the surface of high-density lipoproteins (HDL) impact particle function...