Biography Ernesto Nakayasu is a senior research scientist focused on understanding molecular mechanisms of diseases. Nakayasu has been applying systems biology and mass spectrometry-based omics measurements to study how pathogens and metabolic alterations cause human diseases; the goal is to...
Filter results
Category
- Scientific Discovery (33)
- Biology (32)
- Earth System Science (22)
- Integrative Omics (16)
- Human Health (12)
- Microbiome Science (10)
- Chemical & Biological Signatures Science (1)
- Chemistry (1)
- Computational Research (1)
- Ecosystem Science (1)
- Materials Science (1)
- National Security (1)
- Plant Science (1)
- Weapons of Mass Effect (1)
Tags
- (-) Omics (26)
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (34)
- Mass spectrometry data (32)
- Multi-Omics (32)
- Viruses (28)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- Synthetic (14)
- Genomics (13)
- sequencing (13)
- West Nile virus (13)
- High Throughput Sequencing (11)
- Influenza A (11)
- TA2 (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- TA1 (10)
- Ebola (9)
Last updated on 2023-02-23T19:37:46+00:00 by LN Anderson PerCon SFA Project Publication Experimental Data Catalog The Persistence Control of Engineered Functions in Complex Soil Microbiomes Project (PerCon SFA) at Pacific Northwest National Laboratory ( PNNL ) is a Genomic Sciences Program...
Datasets
3
"Visualizing the Hidden Half: Plant-Microbe Interactions in the Rhizosphere" Plant roots and the associated rhizosphere constitute a dynamic environment that fosters numerous intra- and interkingdom interactions, including metabolite exchange between plants and soil mediated by root exudates and the...
Last updated on 2024-03-21T18:47:20+00:00 by LN Anderson MPLEx Method Protocol for Integrative Single-Sample Processing for Subsequent Proteomic, Metabolomic, and Lipidomic Analyses Method Source Contributions LN Anderson , Data Curation The Metabolite, Protein and Lipid Extraction (MPLEx) method...
Category
"Deconstructing the Soil Microbiome into Reduced-Complexity Functional Modules" The soil microbiome represents one of the most complex microbial communities on the planet, encompassing thousands of taxa and metabolic pathways, rendering holistic analyses computationally intensive and difficult. Here...
Category
The Thermo Scientific™ Q Exactive™ GC hybrid quadrupole Orbitrap Mass Spectrometer provides an unmatched combination of sensitivity, mass-accuracy and resolving power in a single analysis for the highest confidence in compound discovery, identification, and quantitation. With best-in-class...
Category
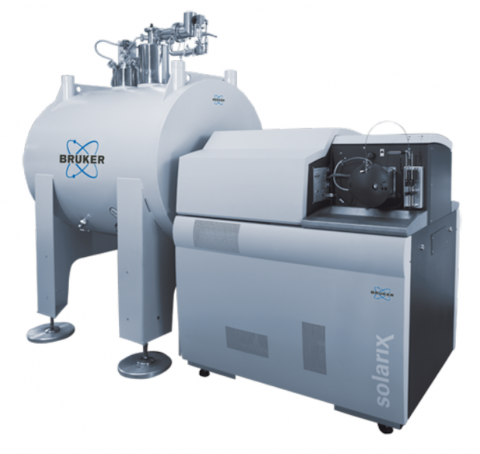
Last updated on 2025-02-03T23:25:06+00:00 by LN Anderson The Bruker Daltonics SolariX Magnetic Resonance Mass Spectrometry (MRMS) instruments are available for different magnetic field strengths of 7T, 12T and 15T. The SolariX XR and 2xR instruments use magnetron control technology and a newly...
Category
Last updated on 2025-02-03T23:25:06+00:00 by LN Anderson The Thermo Scientific™ Velos Pro™ Orbitrap Mass Spectrometer instrument data source combines advanced mass accuracy with an ultra-high resolution Orbitrap mass analyzer for increased sensitivity, enabling molecular weight determination for...
Last updated on 2025-02-03T23:25:06+00:00 by LN Anderson The IONTOF TOF.SIMS 5 data source is a time-of-flight secondary ion mass spectrometer and powerful surface analysis tool used to investigate scientific questions in biological, environmental, and energy research. Among the most sensitive of...
The Nikon Eclipse Ti2-E is a motorized and intelligent model for advanced imaging applications. Compatible with PFS, auto correction collar, and external phase contrast system. The base of choice for live-cell imaging, high-content applications, confocal and super-resolution.As research trends...
Category
The innovative Elstar™ electron column forms the basis of the Helios NanoLab’s outstanding high resolution imaging performance. The Elstar features unique technologies, such as constant power lenses for higher thermal stability, electrostatic scanning for faster, higher accurate imaging, and unique...
Category
Datasets
1
The Illumina HiSeq X System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for high throughput, production-scale sequencing laboratories. Built off the HiSeq 2500 System, harnessing the patterned flow cell technology originally developed...
Last updated on 2025-02-03T23:25:06+00:00 by LN Anderson The Thermo Scientific™ Q Exactive™ Plus Mass Spectrometer benchtop LC-MS/MS system combines quadruple precursor ion selection with high-resolution, accurate-mass (HRAM) Orbitrap detection to deliver exceptional performance and versatility...
Stanford Synchrotron Radiation Lightsource Experimental Station 14-3b is a bending magnet side station dedicated to X-Ray Imaging and Micro X-Ray Absorption Spectroscopy of biological, biomedical, materials, and geological samples. Station 14-3b is equipped with specialized instrumentation for XRF...