Filter results
Content type
Tags
- (-) metagenomics (4)
- (-) Proteomics (4)
- (-) species volatility (2)
- Omics (22)
- Soil Microbiology (21)
- sequencing (13)
- Genomics (12)
- High Throughput Sequencing (9)
- Metagenomics (9)
- PerCon SFA (9)
- Microbiome (8)
- Fungi (6)
- Imaging (6)
- Mass Spectrometry (6)
- Mass Spectrometer (5)
- Sequencer System (5)
- Synthetic Biology (5)
- Microscopy (4)
- Sequencing (4)
- soil microbiology (4)
- Spectroscopy (4)
- Amplicon Sequencing (3)
- Climate Change (3)
- IAREC (3)
- Mass spectrometry data (3)
- metabolomics (3)
- Viruses (3)
- Whole Genome Sequencing (3)
- microbiome stability (2)
- Output Databases (2)
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from KPBS field site in Manhattan, KS. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from COBS field site in Boone County, IA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Complete replicate terabase metagenome (TmG.2.0) of grassland soil microbiome collections from IAREC field site in Prosser, WA. Metagenome (unclassified soil sequencing) Data DOI Package, version 2.0.
Category
Viral communities detected from three large grassland soil metagenomes with historically different precipitation moisture regimes.
Category
Last updated on 2024-03-03T02:26:52+00:00 by LN Anderson The Thermo Scientific™ Velos Pro™ Orbitrap Mass Spectrometer instrument data source combines advanced mass accuracy with an ultra-high resolution Orbitrap mass analyzer for increased sensitivity, enabling molecular weight determination for...
Last updated on 2024-03-03T02:26:52+00:00 by LN Anderson The Thermo Scientific™ Q Exactive™ Plus Mass Spectrometer benchtop LC-MS/MS system combines quadruple precursor ion selection with high-resolution, accurate-mass (HRAM) Orbitrap detection to deliver exceptional performance and versatility...
Bruker Daltonics SolariX Magnetic Resonance Mass Spectrometry (MRMS) instruments are available for different magnetic field strengths of 7T, 12T and 15T. The SolariX XR and 2xR instruments use magnetron control technology and a newly developed, high sensitivity, low noise preamplifier to exploit...
Category
Category
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Metatranscriptomic data from MSC-2. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33232 Metatranscriptomic data from MSC-2 12 fastq files (6 forward read, 6 reverse...
Category
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Category
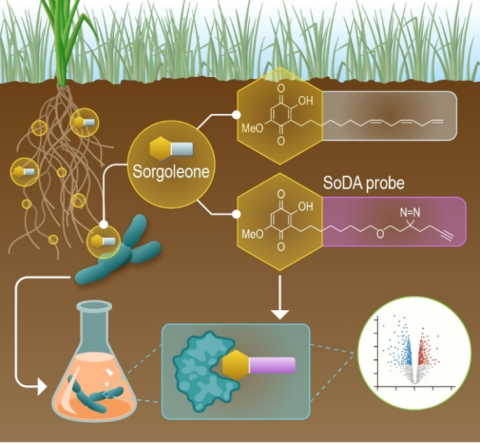
Last updated on 2024-04-19T19:12:08+00:00 by LN Anderson PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...