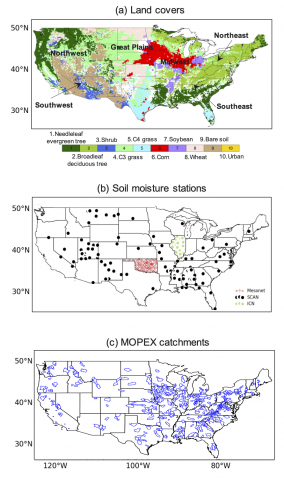
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL008 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome and proteome analysis in human lung 2B4 cells (clonal derivative of Calu-3 cells) infected with either icSARS CoV, icSARS-deltaNSP16 or...
Filter results
Category
- (-) Scientific Discovery (57)
- Biology (52)
- Human Health (51)
- Integrative Omics (42)
- Earth System Science (4)
- Energy Resiliency (4)
- National Security (4)
- Computing & Analytics (3)
- Renewable Energy (3)
- Atmospheric Science (2)
- Computational Research (2)
- Data Analytics & Machine Learning (2)
- Coastal Science (1)
- Computational Mathematics & Statistics (1)
- Cybersecurity (1)
- Data Analytics & Machine Learning (1)
- Distribution (1)
- Electric Grid Modernization (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Grid Analytics (1)
- Grid Cybersecurity (1)
- High-Performance Computing (1)
- Microbiome Science (1)
- Solar Energy (1)
- Visual Analytics (1)
- Wind Energy (1)
Content type
Dataset Type
- (-) Transcriptomics (51)
- (-) Microarray (9)
- (-) Simulation Results (5)
- Omics (83)
- Proteomics (38)
- Sequencing (38)
- Metabolomics (27)
- Lipidomics (26)
- Amplicon (16S, ITS) (14)
- Metagenomics (9)
- Genomics (4)
- Computed Analysis (3)
- ChIP-seq (2)
- Geospacial (1)
- Mass Spectra (1)
- Phenomics (1)
- Staff (1)
- Whole genome sequencing (WGS) (1)
Tags
- Virology (50)
- Gene expression profile data (40)
- Immune Response (40)
- Time Sampled Measurement Datasets (37)
- Differential Expression Analysis (35)
- Homo sapiens (24)
- Mass spectrometry data (18)
- Multi-Omics (17)
- MERS-CoV (14)
- Mus musculus (13)
- West Nile virus (9)
- Health (8)
- Virus (8)
- Viruses (8)
- Ebola (7)
- Influenza A (7)
- Resource Metadata (6)
- Microarray (5)
- Human Interferon (4)
- Response to type I interferon (3)
- Response to type I & type II interferon (2)
- Viral Experiment (2)
- Amplicon Sequencing (1)
- High-Performance Computing (1)
- Host Response (1)
- Machine Learning (1)
- MultiSector Dynamics (1)
- Output Databases (1)
- Secondary Dataset Download (1)
- Synthetic Biology (1)
Systems Virology Lethal Human Virus, SARS-CoV Experiment SHAE003 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome analysis in Human Airway Epithelial (HAE) cells infected with SARS-CoV, SARS deltaORF6, SARS BatSRBD mutants in a longitudinal study...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SHAE004 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome analysis in Human Airway Epithelial (HAE) cells infected with SARS-CoV, SARS deltaORF6, SARS BatSRBD mutants in a longitudinal study...
Category
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WLN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
These GCAM v4.3 SSP-RCP-GCM Output Databases are made available under the Open Data Commons Attribution License: http://opendatacommons.org/licenses/by/1.0/ . GCAM v4.3 SSP-RCP-GCM plausible solution databases. Supplemental dataset to: Graham N.T., M.I. Hejazi, M. Chen, E. Davies, J.A. Edmonds, S.H...
Category
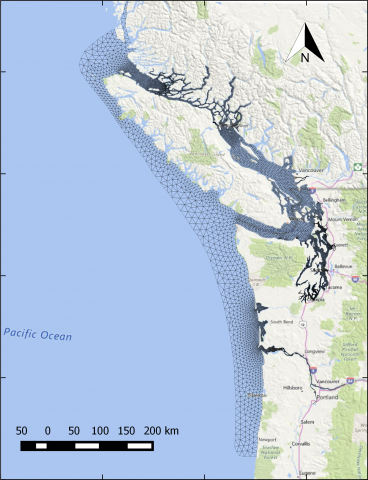
The year 2014 solution files are based on model calibration effort based on inputs and data from Year 2014 as described in Khangaonkar et al. 2018). It includes water surface elevation, currents, temperature and salinity at an hourly interval. These netcdf files with 24 hourly records include...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
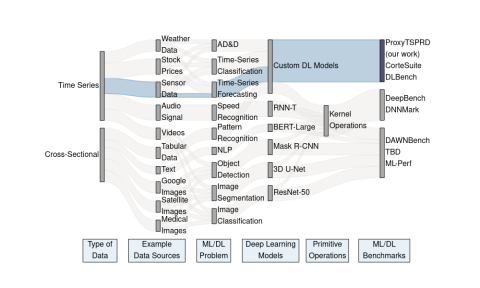
ProxyTSPRD profiles are collected using NVIDIA Nsight Systems version 2020.3.2.6-87e152c and capture computational patterns from training deep learning-based time-series proxy-applications on four different levels: models (Long short-term Memory and Convolutional Neural Network), DL frameworks...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
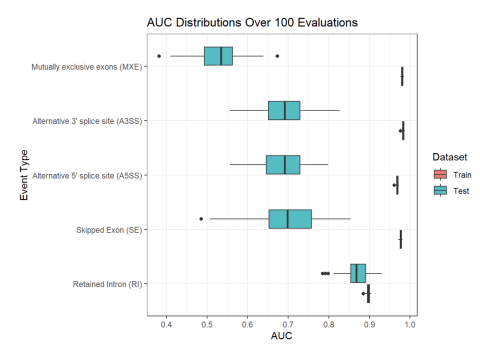
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...