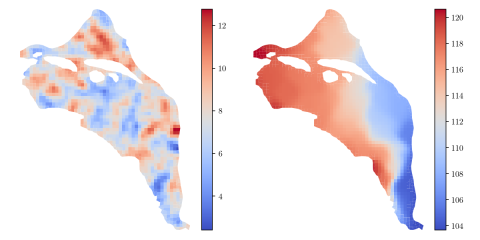
HDF5 file containing 10,000 hydraulic transmissivity inputs and the corresponding hydraulic pressure field outputs for a two-dimensional saturated flow model of the Hanford Site. The inputs are generated by sampling a 1,000-dimensional Kosambi-Karhunen-Loève (KKL) model of the transmissivity field...
Filter results
Category
- Biology (14)
- Scientific Discovery (14)
- Earth System Science (11)
- Human Health (5)
- Microbiome Science (5)
- Computational Research (2)
- Data Analytics & Machine Learning (2)
- Computational Mathematics & Statistics (1)
- Computational Mathematics & Statistics (1)
- Computing & Analytics (1)
- Data Analytics & Machine Learning (1)
- National Security (1)
Content type
Tags
- (-) Metagenomics (7)
- (-) Sequencer System (5)
- Virology (63)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (27)
- Multi-Omics (23)
- Omics (17)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (14)
- Health (13)
- Soil Microbiology (13)
- Virus (13)
- West Nile virus (11)
- Synthetic (10)
- Ebola (9)
- Genomics (9)
- Influenza A (9)
- sequencing (9)
- Resource Metadata (8)
- High Throughput Sequencing (6)
- Microarray (6)
- Mass Spectrometer (5)
- Proteomics (5)
- Imaging (4)
- Microscopy (4)
The Sequel II System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by PacBio , and is designed for high throughput, production-scale sequencing laboratories. Originally released in 2015, the Sequel system provides Single Molecule, Real-Time (SMRT) sequencing core...
The Illumina MiSeq System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for sequencing data acquisition using synthesis technology to provide an end-to-end solution (cluster generation, amplification, sequencing, and data analysis) in a...
The Illumina HiSeq X System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for high throughput, production-scale sequencing laboratories. Built off the HiSeq 2500 System, harnessing the patterned flow cell technology originally developed...
The Illumina HiSeq 4000 System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for high throughput, production-scale sequencing laboratories. Built off the HiSeq 2500 System and harnessing the patterned flow cell technology originally...
The Sequel II System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by PacBio , and is designed for high throughput, production-scale sequencing laboratories. Originally released in 2015, the Sequel system provides Single Molecule, Real-Time (SMRT) sequencing core...
Category
Category
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. IA-TmG.2.0 (Metagenome, IA). [Data Set] PNNL DataHub. https://doi.org/10.25584/IATmG2/1770333 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. WA-TmG.2.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG2/1770324 Soil samples were collected in triplicate in the fall of 2017 across the three grassland...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. KS-TmG.2.0 (Metagenome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KSTmG2/1770332 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
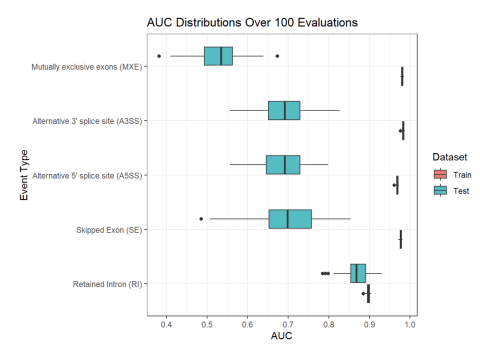
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...