Omics-LHV, West Nile Experiment WCD003 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse dendritic cell response to wild-type West Nile virus (WNV). Overall Design: Mouse dendritic cells (2 x 10^5) were treated with wild-type WNV and collected in parallel...
Filter results
Content type
Tags
- (-) Soil Microbiology (7)
- (-) Virus (3)
- Virology (26)
- Differential Expression Analysis (23)
- Immune Response (23)
- Mass spectrometry data (23)
- Time Sampled Measurement Datasets (23)
- Multi-Omics (22)
- Gene expression profile data (17)
- Homo sapiens (16)
- MERS-CoV (12)
- Mus musculus (7)
- Metagenomics (6)
- sequencing (6)
- Influenza A (5)
- West Nile virus (4)
- Health (3)
- metagenomics (3)
- Microbiome (3)
- Omics (3)
- Sequencing (3)
- soil microbiology (3)
- Viruses (3)
- Ebola (2)
- IAREC (2)
- omics (2)
- Amplicon Sequencing (1)
- Mass Spectrometry (1)
- MPLEX (1)
- Software Data Analysis (1)
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
Category
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL009 New uploads pending The purpose of this SARS experiment was to obtain samples for metabolome and lipidome analysis comparing wild type virus (icSARS) infected Human lung tissue 2B4 (Calu-3 clonal derivative) cells to mock infected cells...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL012 New uploads pending The purpose of this SARS experiment was to obtain samples for metabolome and lipidome analysis comparing wild type virus (icSARS) vs icSARS CoV deltaORF6 infected Human lung tissue 2B4 (Calu-3 clonal derivative)...
Category
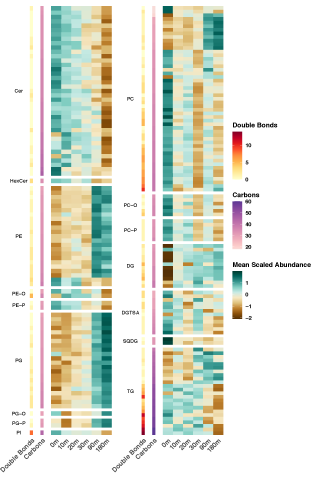
Rapid remodeling of the soil lipidome in response to a drying-rewetting event - Multi-Omics Data Package DOI Data package contents reported here are the first version and contain pre- and post-processed data acquisition and subsequent downstream analysis files using various data source instrument...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. WA-TmG.2.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG2/1770324 Soil samples were collected in triplicate in the fall of 2017 across the three grassland...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. KS-TmG.2.0 (Metagenome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KSTmG2/1770332 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. IA-TmG.2.0 (Metagenome, IA). [Data Set] PNNL DataHub. https://doi.org/10.25584/IATmG2/1770333 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...