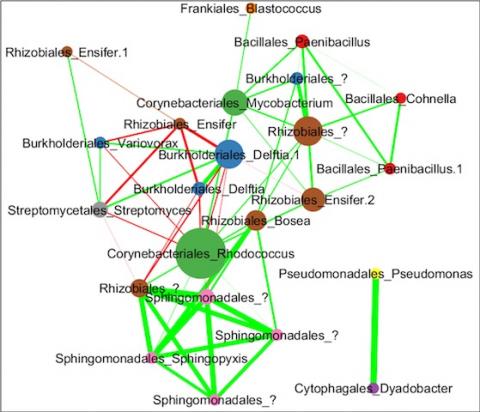
Please cite as : Anderson L.N., J.E. McDermott, and R.S. McClure. 2020. WA-IsoC_MSC1.1.0 (Amplicon 16S rRNA, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WAIsoCMSC1/1635272 The soil microbiome is central to the cycling of carbon and other nutrients and to the promotion of plant growth...
Filter results
Category
- (-) Biology (285)
- (-) Coastal Science (4)
- Scientific Discovery (401)
- Earth System Science (166)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (13)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Renewable Energy (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
- Water Power (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (34)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (25)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- TA1 (9)
- Microbiome (8)
- Post-Translational Modifications (8)
Category
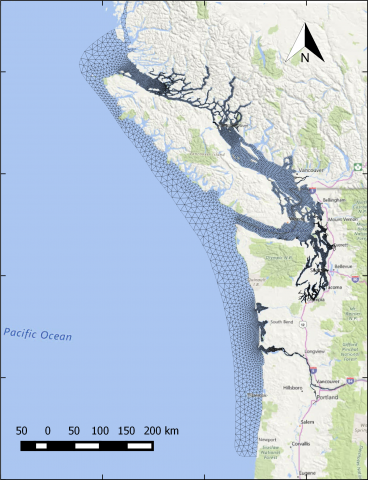
The year 2014 solution files are based on model calibration effort based on inputs and data from Year 2014 as described in Khangaonkar et al. 2018). It includes water surface elevation, currents, temperature and salinity at an hourly interval. These netcdf files with 24 hourly records include...
Category
Category
Category
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
The year 2014 solution files are based on model calibration effort based on inputs and data from Year 2014 as described in Khangaonkar et al. 2018). It includes water surface elevation, currents, temperature and salinity at an hourly interval. These netcdf files with 24 hourly records include...
Category
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WLN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCT001 The purpose of this experiment was to evaluate the host responseto West Nile virus (WNV-NY99) wild-type (strain 382) and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCB001 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM103 The purpose of this experiment was to evaluate the host response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004 (VN1203) virus, mutant VN1203-NS1trunc124, and mock...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM102 The purpose of this experiment was to evaluate the mouse host response to Influenza A virus (subtype H7N9) wild-type strain Influenza A/Anhui/1/2013 (AH1-WT) virus and mutants NS1-103F/106M (AH1-F/M) and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM101 The purpose of this experiment was to evaluate the host mouse response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004, Influenza A/Vietnam/1203/2004 mutant strains PB2-627E...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL104 The purpose of this experiment was to evaluate the host response to pandemic Influenza A virus (subtype H1N1) , natural isolate Influenza A/California/04/2009 virus. Sample data was obtained from human lung...