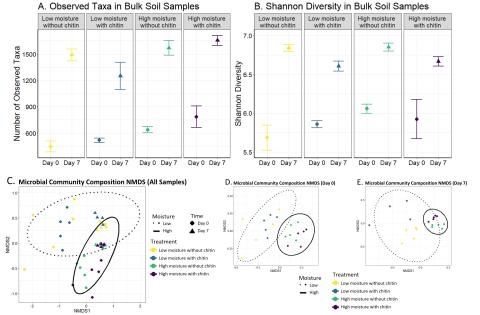
Please cite as : Nicholas J Reichart, Sheryl Bell, Vanessa A. Garayburu-Caruso, Natalie Sadler, Sharon Zhao, Kirsten Hofmockel. Chitin Decomposition 16S rRNA gene fastq. 2024 . [Data Set] PNNL DataHub. 10.25584/2475041. This data is published under a CC0 license . The authors encourage data reuse...
Filter results
Category
- (-) Biology (258)
- (-) Materials Science (7)
- Scientific Discovery (369)
- Earth System Science (161)
- Human Health (112)
- Integrative Omics (73)
- Microbiome Science (47)
- National Security (31)
- Computational Research (25)
- Computing & Analytics (17)
- Chemical & Biological Signatures Science (12)
- Energy Resiliency (12)
- Weapons of Mass Effect (12)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Atmospheric Science (6)
- Data Analytics & Machine Learning (6)
- Renewable Energy (6)
- Visual Analytics (6)
- Coastal Science (4)
- Ecosystem Science (4)
- Energy Storage (3)
- Plant Science (3)
- Solar Energy (3)
- Bioenergy Technologies (2)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Grid Cybersecurity (2)
- Transportation (2)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Wind Energy (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (34)
- Mass spectrometry data (32)
- Multi-Omics (32)
- Viruses (27)
- Omics (25)
- Health (23)
- Mass Spectrometry (23)
- Soil Microbiology (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- TA2 (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- TA1 (10)
- Ebola (9)
- Microbiome (8)
Created on 2024-10-16T17:27:40+00:00 by LN Anderson and is pending updates.
Created on 2025-06-10T00:34:54+00:00 by LN Anderson
Created on 2024-10-16T17:27:40+00:00 by LN Anderson . Pending updates will be available for public release before or by 2025-10-01.
Created on 2024-10-16T17:27:40+00:00 by LN Anderson . Pending updates will be available for public release before or by 2025-10-01.
Created on 2024-10-15T19:55:00+00:00 by LN Anderson ; Last updated 2024-11-13T15:01:44+00:00 and is pending public release before or by 2025-10-01. Human Liver Epithelium Response to HCoV-229E Infection Epigenomics (ACS-DP4) The purpose of this experiment was to evaluate how wild-type Human...
Category
Created on 2024-10-15T19:55:00+00:00 by LN Anderson ; Last updated 2024-11-13T15:01:44+00:00 and is pending public release before or by 2025-10-01. Human Primary Airway Epithelium Response to HCoV-229E Infection Transcriptomics (ACS-DP3) The purpose of this experiment was to evaluate the human host...
Category
Created on 2024-10-14T20:33:54+00:00 by LN Anderson pending updates.
Category
Created on 2024-10-14T20:33:54+00:00 by LN Anderson . Pending updates will be available for public release before or by 2025-10-01.
Category
Created on 2024-10-14T20:33:54+00:00 by LN Anderson ; Last Updated 2024-11-13T15:01:44+00:00 and is pending public release. S. elongatus PCC 7942 Carbon Metabolism Proteomics (MC-DP2) The purpose of this experiment was to understand the regulatory dynamics of carbon fixation in S. elongatus PCC 7942...
Category
Created on updated 2024-10-01T21:34:23+00:00 by LN Anderson ; Last updated 2024-11-13T15:01:44+00:00 and is pending public release. Human Host Cellular Response to HCoV-229E Infection Proteomics (ACS-JM-DP2) The purpose of this experiment was to evaluate the human host cellular response to wild-type...
Created 2024-09-17T19:19:38+00:00 by LN Anderson ; Last updated 2024-10-17T17:30:32+00:00 and is pending public release. Human A549 and MRC5 Cell Response to HCoV-229E Infection Transcriptomics (ACS-DP1) The purpose of this experiment was to evaluate the human host cellular response to wild-type...
Category
Last updated on 2025-01-31T00:37:40+00:00 by LN Anderson High-density Lipoprotein (HDL) Structure and Function (JM-DP1) The purpose of this experiment was to investigate how the interactions between APOA1 and APOA2 on the surface of high-density lipoproteins (HDL) impact particle function...
Category
Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...
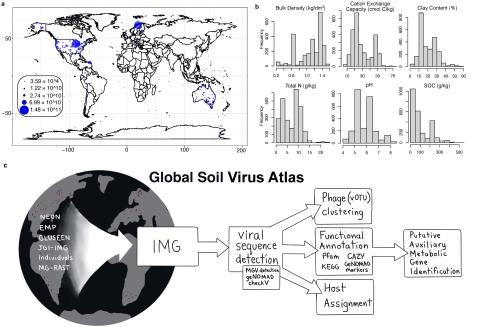
Please cite as : Graham E.B., A. Camargo, R. Wu, R.Y. Neches, M. Nolan, W.C. Nelson, and M.M. Pleake, et al. 2024. Global Soil Virus (GSV) Atlas. [Data Set] PNNL DataHub. 10.25584/2229733 This data is published under a CC0 license . The authors encourage data reuse and request attribution by...