Category
Description
Background
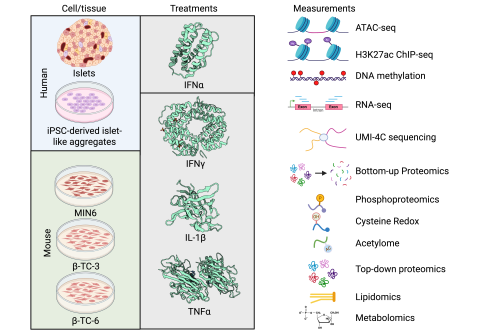
In type 1 diabetes (T1D), autoimmune response and inflammation cause the death of pancreatic β cells, leading to the body’s inability to produce insulin and maintain glucose homeostasis. This process is at least in part mediated by pro-inflammatory cytokines, such as interferon (IFN)α, IFNγ, interleukin (IL)-1β, and tumor necrosis factor (TNF)α, which induce β-cell dysfunction and apoptosis. A deep understanding of the β-cell signaling and regulatory networks induced by these cytokines could lead to the identification of therapeutic targets to prevent T1D development. To study cytokine-mediated islets/β-cell signaling and regulatory networks, a variety of omics experiments have been conducted, including transcriptomics, epigenomics (DNA methylation, UMI-4C, ATAC-seq & ChIP-seq), proteomics (bottom-up, top-down, post-translational modification analysis), lipidomics, and metabolomics.
Impact
The combination of these datasets will be instrumental in identifying signaling components and regulatory factors involved in β-cell stress/death.
Resource Description
This is a compendium of datasets from previously published multi-omics studies of pancreatic islets and β-cell models exposed to pro-inflammatory cytokines. The omics datasets are aggregated and quality-controlled into unified “data packages” for sharing via PNNL DataHub. The resource currently comprises 44 datasets from 19 studies organized into 21 data packages (Pck001–Pck021) spanning data from primary human islets, human β-cell lines (EndoC-βH1, iPSC-derived islet-like cells), and mouse β-cell lines (MIN6, β-TC-3, β-TC-6).
Funding Acknowledgment
This work was supported by the Catalyst Award from the Human Islet Research Network (HIRN) (to E.S.N) (via U24 DK104162) and by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01 DK138335 (to ESN, BJWR, and TOM), U01 DK127505 (to LS and ESN), U01 DK127786 (to RGM, DLE, and TOM), R01 DK060581 (to RGM), R01 DK105588 (to RGM), and P30 DK020595 (to RGM). This work was also supported by Spanish Ministry of Science and Innovation PID2023-151556OB-I00, CNS2024-154742, “la Caixa” Foundation, LCF-PR-HR24-00150 (to LP) and the 2024 EFSD/Lilly Young Investigator Research Award (MR-R), Breakthrough T1D (3-SRA-2023-1379-S-B, 3-IND-2024-1549-I-X, 2-SRA-2024-1524-S-B and IDDP-InSphero 201310390), the Leona M. & Harry B. Helmsley Charitable Trust (Grants #2402-08172 and 2402-08172) and the Win4Excellence – Gene therapy in Wallonie (GT4Health_20230920) (to DLE). FS was supported by JDRF 5-CDA-2022-1176-A-N and (CDA)/3-CDA-2025-1692-A-N.
Citation Policy
To promote discovery, reproducibility, and reuse of NIH-funded project dataset citations, all reuse of project data and metadata download materials should acknowledge all relevant primary and secondary dataset citations, as well as direct corresponding journal articles where permissible. These practices should follow the FORCE11 Joint Declaration of Data Citation Principles and comply with NIH acknowledgement requirements.