Filter results
Category
- (-) Scientific Discovery (194)
- Biology (121)
- Earth System Science (102)
- Human Health (75)
- Integrative Omics (59)
- Microbiome Science (22)
- National Security (14)
- Computing & Analytics (10)
- Energy Resiliency (7)
- Computational Research (6)
- Visual Analytics (6)
- Atmospheric Science (3)
- Coastal Science (3)
- Computational Mathematics & Statistics (3)
- Data Analytics & Machine Learning (3)
- Data Analytics & Machine Learning (3)
- Ecosystem Science (3)
- Renewable Energy (3)
- Chemical & Biological Signatures Science (2)
- Chemistry (2)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Materials Science (2)
- Weapons of Mass Effect (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Energy Efficiency (1)
- Energy Storage (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Plant Science (1)
- Solar Energy (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Dataset Type
- Omics (83)
- Transcriptomics (51)
- Proteomics (38)
- Sequencing (38)
- Metabolomics (27)
- Lipidomics (26)
- Amplicon (16S, ITS) (14)
- Metagenomics (9)
- Microarray (9)
- Simulation Results (5)
- Genomics (4)
- Computed Analysis (3)
- ChIP-seq (2)
- Geospacial (1)
- Mass Spectra (1)
- Phenomics (1)
- Staff (1)
- Whole genome sequencing (WGS) (1)
Tags
- Virology (63)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (27)
- Multi-Omics (23)
- Omics (17)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (14)
- Health (13)
- Soil Microbiology (13)
- Virus (13)
- West Nile virus (11)
- Ebola (9)
- Genomics (9)
- Influenza A (9)
- sequencing (9)
- Resource Metadata (8)
- Metagenomics (7)
- High Throughput Sequencing (6)
- Microarray (6)
- Mass Spectrometer (5)
- Proteomics (5)
- Sequencer System (5)
- Imaging (4)
- Microbiome (4)
- Microscopy (4)
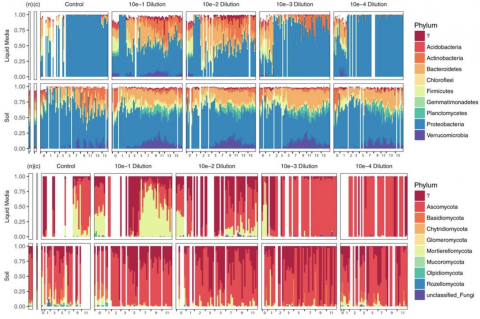
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL008 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome and proteome analysis in human lung 2B4 cells (clonal derivative of Calu-3 cells) infected with either icSARS CoV, icSARS-deltaNSP16 or...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL009 New uploads pending The purpose of this SARS experiment was to obtain samples for metabolome and lipidome analysis comparing wild type virus (icSARS) infected Human lung tissue 2B4 (Calu-3 clonal derivative) cells to mock infected cells...
Category
Category
Category
Category
Category
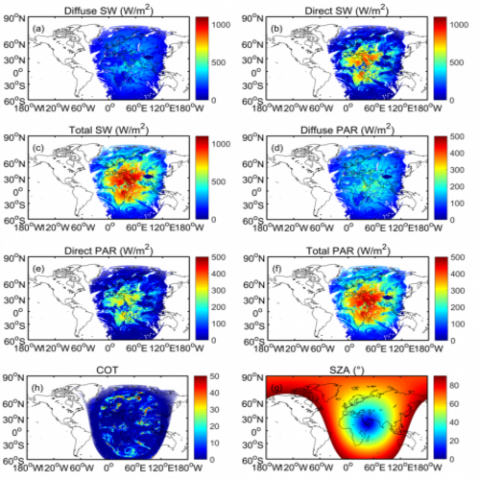
Comprehensive assessment of climate datasets created by statistical or dynamical models is important for effectively communicating model projection and associated uncertainty to stakeholders and decision-makers. The Department of Energy FACETS project aims to foster such communication through...
Category
Category
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WGCN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WSE001 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 virus infection. Sample data was obtained from mouse (strain C57BL6/JAX) blood serum...
Category
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain C57BL...