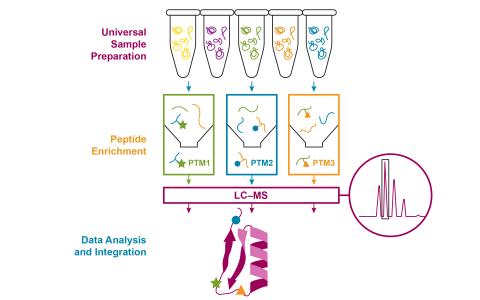
Created 2025-06-03T23:16:59+00:00 by LN Anderson Integrative SP3 Workflow for Multi-PTM Proteomics Profiling (PPI-TZ-DP0) The purpose of this experiment was to evaluate a new standardized method approach for proteomics sample processing designed to facilitate multiplexed quantification of protein...
Filter results
Category
- (-) Integrative Omics (73)
- (-) Microbiome Science (47)
- Scientific Discovery (369)
- Biology (258)
- Earth System Science (161)
- Human Health (112)
- National Security (31)
- Computational Research (25)
- Computing & Analytics (17)
- Chemical & Biological Signatures Science (12)
- Energy Resiliency (12)
- Weapons of Mass Effect (12)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Materials Science (7)
- Atmospheric Science (6)
- Data Analytics & Machine Learning (6)
- Renewable Energy (6)
- Visual Analytics (6)
- Coastal Science (4)
- Ecosystem Science (4)
- Energy Storage (3)
- Plant Science (3)
- Solar Energy (3)
- Bioenergy Technologies (2)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Grid Cybersecurity (2)
- Transportation (2)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Wind Energy (1)
Tags
- Virology (55)
- Immune Response (51)
- Time Sampled Measurement Datasets (50)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (31)
- Mus musculus (19)
- MERS-CoV (18)
- Omics (18)
- West Nile virus (13)
- Influenza A (11)
- Mass Spectrometry (11)
- Ebola (9)
- PerCon SFA (9)
- High Throughput Sequencing (8)
- Genomics (7)
- Proteomics (7)
- Resource Metadata (7)
- Human Interferon (6)
- Microarray (6)
- Omics-LHV Project (6)
- Imaging (5)
- Sequencer System (5)
- Synthetic Biology (5)
- Mass Spectrometer (4)
- metabolomics (4)
- Soil Microbiology (4)
- Microscopy (3)
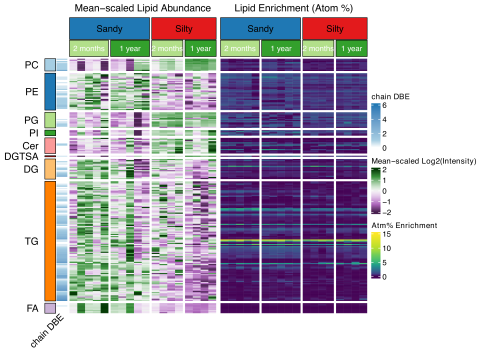
Data package for Lipids represent a dynamic, yet stable pool of microbially-derived soil carbon This data is published under a CC0 license . The authors encourage data reuse and request attribution by referencing the below citations for the data packages and associated manuscript. Please cite as...
Please cite as : Rodríguez-Ramos, Josué, Natalie Sadler, Elias K. Zegeye, Yuliya Farris, Samuel Purvine, Sneha Couvillion, William C. Nelson, and Kirsten Hofmockel. Environmental matrix and moisture are key determinants of microbial phenotypes expressed in a reduced complexity soil-analog. [Data Set...
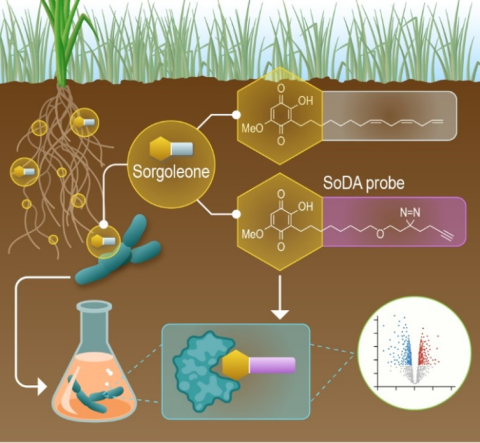
Last updated on 2024-09-17T15:49:08+00:00 by LN Anderson PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
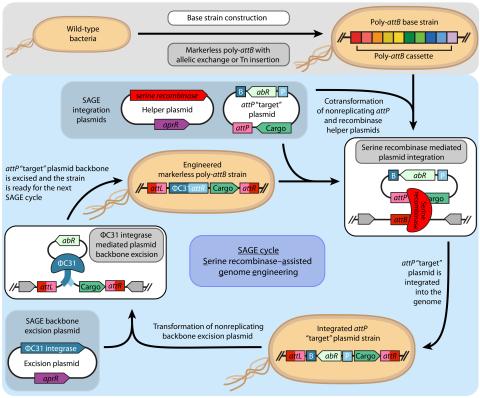
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
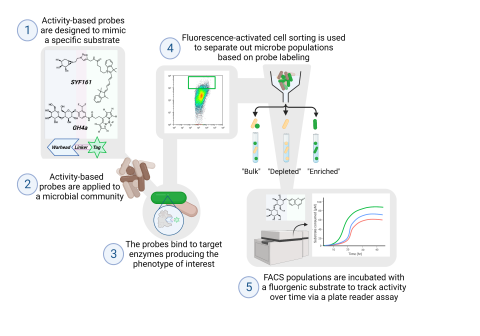
Last updated on 2024-10-04T19:31:35+00:00 by LN Anderson Activity-Based Probe Fluorescence-Activated Cell Sorting (ABP-FACS) of Soil Microbes (NR-DP0) Soil samples were incubated with either 0.2% cellobiose for three days or 0.2% xylan for nine days. Microbes were extracted from the soil matrix...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
Omics-LHV, West Nile Experiment WGCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in primary mouse granule neuron cells infected with wild type West Nile virus (WNV-NY99 clone 382, WNVWT) and mutant virus (WNVE218A). Overall Design: Granule cell neurons from...
Category
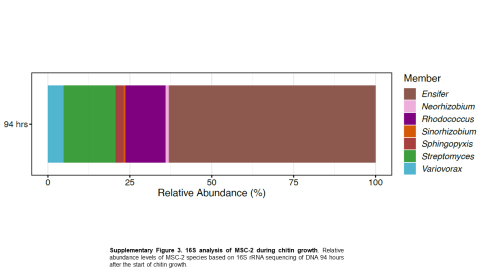
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Category
This data set provides the ITS fungal community composition via DNA and cDNA sequence analysis for peat and sand ingrowth cores collected before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE)...
Category
This data set provides the 16S microbial community composition via DNA sequence analysis from ingrowth peat and sand cores at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the...
Category
This data set provides ITS fungal community composition via DNA and cDNA sequence analysis at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a Qiagen...
Category
Last updated on 2025-01-16T22:12:33+00:00 by LN Anderson Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68" Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for...