Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
Filter results
Category
- (-) Biology (287)
- Scientific Discovery (404)
- Earth System Science (167)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (25)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
- Post-Translational Modifications (8)
- TA1 (8)
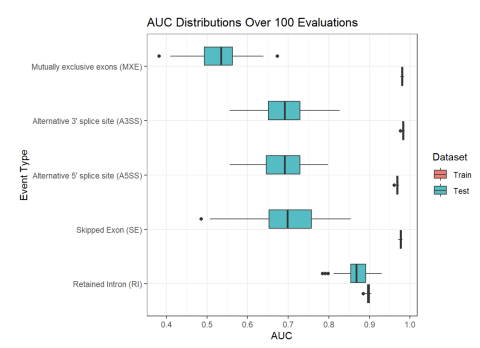
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
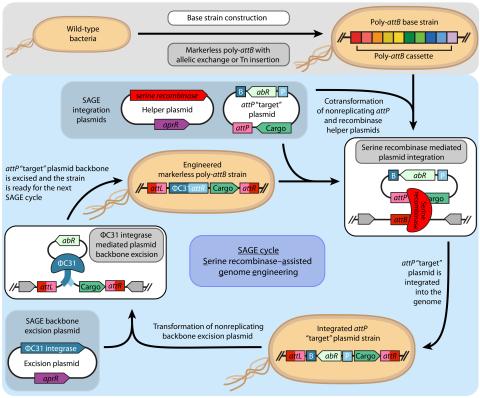
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
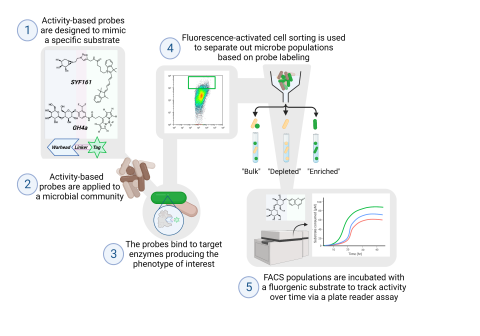
Last updated on 2025-08-11 by LN Anderson Activity-Based Probe Fluorescence-Activated Cell Sorting (ABP-FACS) of Soil Microbes (NR-DP0) Soil samples were incubated with either 0.2% cellobiose for three days or 0.2% xylan for nine days. Microbes were extracted from the soil matrix, incubated...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
Omics-LHV, West Nile Experiment WCD003 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse dendritic cell response to wild-type West Nile virus (WNV). Overall Design: Mouse dendritic cells (2 x 10^5) were treated with wild-type WNV and collected in parallel...
Category
Omics-LHV, West Nile Experiment WCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse cerebral cortex neurons in response to wild-type West Nile Virus (WNV; WNV-NY99 382) and mutant WNV-E218A (WNV-NY99 382) viral infection. Overall Design: Mouse cortical...
Category
Omics-LHV, West Nile Experiment WGCN004 The purpose of this West Nile experiment was to obtain samples for omics analysis in primary mouse granule neuron cells infected with wild type West Nile virus (WNV-NY99 clone 382, WNVWT) and mutant virus (WNVE218A). Overall Design: Granule cell neurons from...
Category
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
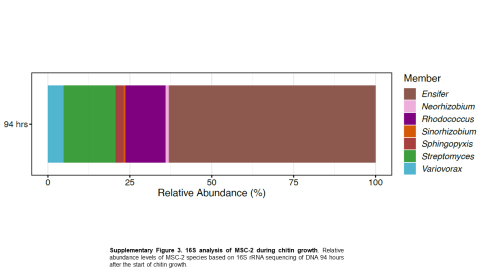
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Category
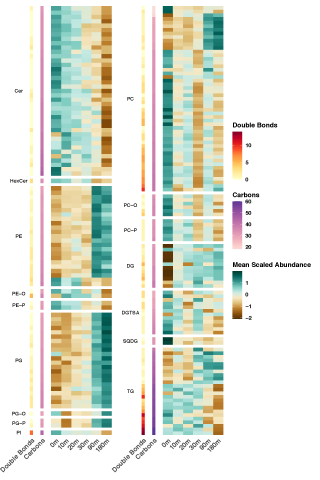
Rapid remodeling of the soil lipidome in response to a drying-rewetting event - Multi-Omics Data Package DOI Data package contents reported here are the first version and contain pre- and post-processed data acquisition and subsequent downstream analysis files using various data source instrument...
Category
This data set provides ITS fungal community composition via DNA and cDNA sequence analysis at the time of peat coring for Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2014-2017 from the Spruce and Peatlands Under Changing Environments (SPRUCE). Samples were extracted using a Qiagen...
Category
This data set provides the 16S microbial community composition via DNA sequence analysis from ingrowth peat and sand cores at the South End bog in 2013. These samples were collected outside the experimental enclosures and are pre-treatment with no experimental manipulation. These are part of the...
Category
This data set provides the ITS fungal community composition via DNA and cDNA sequence analysis for peat and sand ingrowth cores collected before and during Deep Peat Heating (DPH) and Whole Ecosystem Warming (WEW) for 2015-2016 from the Spruce and Peatlands Under Changing Environments (SPRUCE)...