Last updated on 2024-03-21T18:47:20+00:00 by LN Anderson MPLEx Method Protocol for Integrative Single-Sample Processing for Subsequent Proteomic, Metabolomic, and Lipidomic Analyses Method Source Contributions LN Anderson , Data Curation The Metabolite, Protein and Lipid Extraction (MPLEx) method...
Filter results
Category
- (-) Scientific Discovery (307)
- Biology (198)
- Earth System Science (136)
- Human Health (102)
- Integrative Omics (73)
- Microbiome Science (42)
- Computational Research (23)
- National Security (21)
- Computing & Analytics (14)
- Chemistry (10)
- Energy Resiliency (9)
- Data Analytics & Machine Learning (8)
- Materials Science (7)
- Visual Analytics (6)
- Chemical & Biological Signatures Science (5)
- Computational Mathematics & Statistics (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Coastal Science (4)
- Ecosystem Science (4)
- Renewable Energy (4)
- Data Analytics & Machine Learning (3)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Energy Storage (2)
- Grid Cybersecurity (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Viruses (26)
- Omics (25)
- Health (23)
- Virus (23)
- Soil Microbiology (21)
- MERS-CoV (18)
- Mus musculus (18)
- Mass Spectrometry (14)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- Ebola (11)
- Influenza A (11)
- PerCon SFA (10)
- High Throughput Sequencing (9)
- Metagenomics (9)
- Resource Metadata (9)
- Microbiome (8)
- Proteomics (8)
- Microarray (7)
- Synthetic Biology (7)
- Imaging (6)
Biography Carrie Nicora is a senior research scientist (chemist III) at PNNL, specializing in high-throughput scientific research sample management using modular automation techniques for processing a wide range of biological specimens for proteomic, metabolomic, and lipidomic analysis. She is an...
Category
Biography Dr. Smith's research interests span the development of advanced analytical methods and instrumentation, with particular emphasis on high-resolution separations and mass spectrometry, and their applications in biological and biomedical research. This has included creating and applying new...
Biography Kelly is a senior data scientist in the Computational Biology group within the Biological Sciences Division at Pacific Northwest National Laboratory (PNNL). After earning a MS in Biostatistics from the University of Washington in 2012, she worked at a cancer research company for two years...
David Degnan is a biological data scientist who develops bioinformatic and statistical pipelines for multi-omics data, specifically the fields of proteomics, metabolomics, and multi-omics (phenotypic) data integration. He has experience with top-down & bottom-up proteomics analysis, genomics &...
The Illumina MiSeq System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for sequencing data acquisition using synthesis technology to provide an end-to-end solution (cluster generation, amplification, sequencing, and data analysis) in a...
Last updated on 2024-03-03T02:26:52+00:00 by LN Anderson The Thermo Scientific™ Q Exactive™ Plus Mass Spectrometer benchtop LC-MS/MS system combines quadruple precursor ion selection with high-resolution, accurate-mass (HRAM) Orbitrap detection to deliver exceptional performance and versatility...
Rigaku Rapid II Microbeam is one of the most versatile micro-diffraction XRD system in materials analysis, using advanced imaging plate technology for measuring diffraction patterns and diffuse scattering from a wide range of materials. The RAPID™ II Curved Detector X-Ray Diffraction (XRD) System's...
Category
The Illumina HiSeq 4000 System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for high throughput, production-scale sequencing laboratories. Built off the HiSeq 2500 System and harnessing the patterned flow cell technology originally...
The srpAnalytics modeling pipeline for processing new data for the OSU-PNNL Superfund Research Program Analytics Portal located at https://github.com/sgosline/srpAnalytics Data License MIT License Copyright (c) 2020 Sara JC Gosline . Permission is hereby granted, free of charge, to any person...
Category
The Illumina HiSeq X System Sequencer is a high-throughput DNA sequencer machine developed and manufactured by Illumina , and is designed for high throughput, production-scale sequencing laboratories. Built off the HiSeq 2500 System, harnessing the patterned flow cell technology originally developed...
Last updated on 2024-03-03T02:26:52+00:00 by LN Anderson The Thermo Scientific™ Velos Pro™ Orbitrap Mass Spectrometer instrument data source combines advanced mass accuracy with an ultra-high resolution Orbitrap mass analyzer for increased sensitivity, enabling molecular weight determination for...
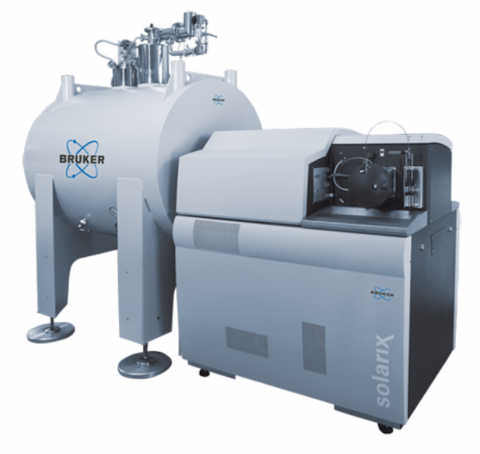
Bruker Daltonics SolariX Magnetic Resonance Mass Spectrometry (MRMS) instruments are available for different magnetic field strengths of 7T, 12T and 15T. The SolariX XR and 2xR instruments use magnetron control technology and a newly developed, high sensitivity, low noise preamplifier to exploit...
Category
The IONTOF TOF.SIMS 5 data source is a time-of-flight secondary ion mass spectrometer and powerful surface analysis tool used to investigate scientific questions in biological, environmental, and energy research. Among the most sensitive of surface analysis tools, it uses a high-vacuum technique...
The Nikon Eclipse Ti2-E is a motorized and intelligent model for advanced imaging applications. Compatible with PFS, auto correction collar, and external phase contrast system. The base of choice for live-cell imaging, high-content applications, confocal and super-resolution.As research trends...