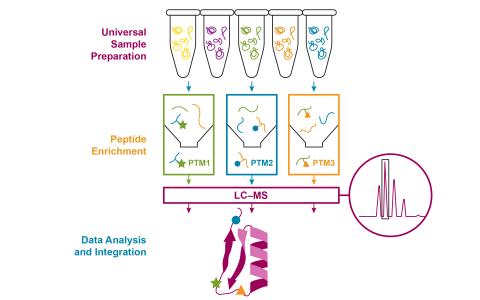
Created 2025-06-03T23:16:59+00:00 by LN Anderson Integrative SP3 Workflow for Multi-PTM Proteomics Profiling (PPI-TZ-DP0) The purpose of this experiment was to evaluate a new standardized method approach for proteomics sample processing designed to facilitate multiplexed quantification of protein...
Filter results
Content type
Tags
- (-) Predictive Phenomics (34)
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Mass spectrometry data (32)
- Multi-Omics (32)
- Viruses (28)
- Omics (26)
- Mass Spectrometry (24)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- Synthetic (14)
- Genomics (13)
- sequencing (13)
- West Nile virus (13)
- High Throughput Sequencing (11)
- Influenza A (11)
- TA2 (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- TA1 (10)
- Ebola (9)
Created 2024-12-22T20:31:04+00:00 by LN Anderson ; Last updated 2025-02-04T20:14:18+00:00 and is pending public release. Rhodotorula toruloides Nitrogen Limitation PTM Profiling Multi-Omics (TZ-DP1) The purpose of this experiment was to evaluate the regulatory stress response of Oleaginous yeast...
Category
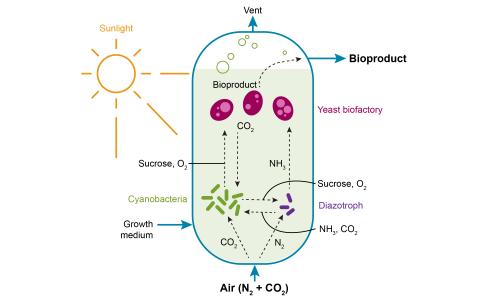
Created 2024-12-20T23:47:47+00:00 by LN Anderson ; Last updated 2025-05-09T15:55:34+00:00 S. elongatus PCC 7942 Circadian Control Bioproduction Metabolomics (PB-DP5) The purpose of this experiment was to evaluate how circadian clock regulation impacts carbon partitioning between storage, growth, and...
Category
Created 2024-12-20T23:47:47+00:00 by LN Anderson ; Last updated 2025-05-09T15:55:34+00:00 S. elongatus PCC 7942 Circadian Control Bioproduction Transcriptomics (PB-DP3) The purpose of this experiment was to evaluate how circadian clock regulation impacts carbon partitioning between storage, growth...
Category
Created on 2024-10-16T17:27:40+00:00 by LN Anderson . Pending updates will be available for public release before or by 2025-10-01.
Created on 2024-10-16T17:27:40+00:00 by LN Anderson and is pending updates.
Created on 2025-06-10T00:34:54+00:00 by LN Anderson
Created on 2024-10-16T17:27:40+00:00 by LN Anderson . Pending updates will be available for public release before or by 2025-10-01.
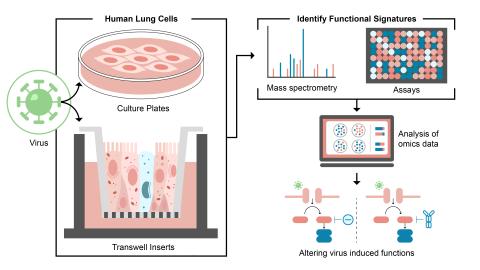
Created on 2024-10-15T19:55:00+00:00 by LN Anderson ; Last updated 2024-11-13T15:01:44+00:00 and is pending public release before or by 2025-10-01. Human Liver Epithelium Response to HCoV-229E Infection Epigenomics (ACS-DP4) The purpose of this experiment was to evaluate how wild-type Human...
Category
Created on 2024-10-14T20:33:54+00:00 by LN Anderson pending updates.
Category
Created on 2024-10-15T19:55:00+00:00 by LN Anderson ; Last updated 2024-11-13T15:01:44+00:00 and is pending public release before or by 2025-10-01. Human Primary Airway Epithelium Response to HCoV-229E Infection Transcriptomics (ACS-DP3) The purpose of this experiment was to evaluate the human host...
Category
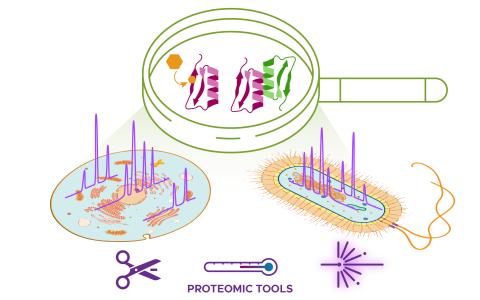
Created on 2024-10-14T20:33:54+00:00 by LN Anderson ; Last Updated 2024-11-13T15:01:44+00:00 and is pending public release. S. elongatus PCC 7942 Carbon Metabolism Proteomics (MC-DP2) The purpose of this experiment was to understand the regulatory dynamics of carbon fixation in S. elongatus PCC 7942...
Category
Created on 2024-10-14T20:33:54+00:00 by LN Anderson . Pending updates will be available for public release before or by 2025-10-01.
Category
Created on updated 2024-10-01T21:34:23+00:00 by LN Anderson ; Last updated 2024-11-13T15:01:44+00:00 and is pending public release. Human Host Cellular Response to HCoV-229E Infection Proteomics (ACS-JM-DP2) The purpose of this experiment was to evaluate the human host cellular response to wild-type...
Created 2024-09-17T19:19:38+00:00 by LN Anderson ; Last updated 2024-10-17T17:30:32+00:00 and is pending public release. Human A549 and MRC5 Cell Response to HCoV-229E Infection Transcriptomics (ACS-DP1) The purpose of this experiment was to evaluate the human host cellular response to wild-type...