Filter results
Category
- (-) Earth System Science (136)
- (-) Integrative Omics (74)
- (-) Computational Research (25)
- (-) Grid Cybersecurity (2)
- Scientific Discovery (309)
- Biology (200)
- Human Health (104)
- Microbiome Science (42)
- National Security (22)
- Computing & Analytics (15)
- Chemistry (10)
- Energy Resiliency (10)
- Data Analytics & Machine Learning (9)
- Computational Mathematics & Statistics (7)
- Materials Science (7)
- Visual Analytics (6)
- Chemical & Biological Signatures Science (5)
- Renewable Energy (5)
- Weapons of Mass Effect (5)
- Atmospheric Science (4)
- Coastal Science (4)
- Data Analytics & Machine Learning (4)
- Ecosystem Science (4)
- Plant Science (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Energy Efficiency (2)
- Energy Storage (2)
- Solar Energy (2)
- Bioenergy Technologies (1)
- Computational Mathematics & Statistics (1)
- Grid Analytics (1)
- High-Performance Computing (1)
- Subsurface Science (1)
- Terrestrial Aquatics (1)
- Transportation (1)
- Wind Energy (1)
Tags
- Virology (57)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (30)
- Multi-Omics (30)
- Omics (24)
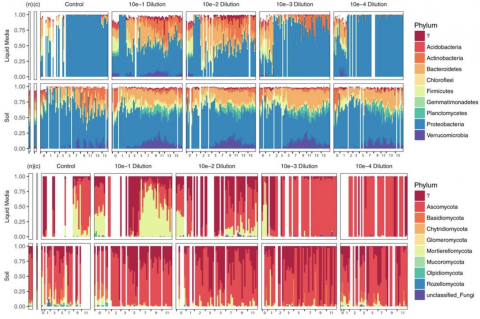
- Soil Microbiology (21)
- MERS-CoV (18)
- Mus musculus (18)
- Mass Spectrometry (13)
- sequencing (13)
- West Nile virus (13)
- Ebola (11)
- Influenza A (11)
- Genomics (10)
- Metagenomics (9)
- Resource Metadata (9)
- Microbiome (8)
- Fungi (6)
- High Throughput Sequencing (6)
- Human Interferon (6)
- Imaging (6)
- Microarray (6)
- Omics-LHV Project (6)
- Type 1 Diabetes (6)
- Mass Spectrometer (5)
- Proteomics (5)
Category
Category
Category
Category
Category
Category
Category
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL003 The purpose of this experiment was to evaluate the human host response to wild-type infectious clone of Middle Eastern Respiratory Syndrome coronavirus (icMERS-CoV) infection and mockulum. Sample data was obtained...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WLN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WDC010 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from primary mouse...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WDC011 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from primary mouse...