Influenza A Experiment IM104 Processed Omics Data Unavailable This Influenza experiment evaluated mouse lung expression after etoposide treatment and infection with a pandemic H1N1 influenza strain. Related Experimental Data BioProject: PRJNA382278 GEO: GSE97555 (mRNA transcriptome response)...
Filter results
Tags
- (-) Influenza A (9)
- (-) Genomics (4)
- Virology (64)
- Immune Response (48)
- Time Sampled Measurement Datasets (45)
- Gene expression profile data (42)
- Differential Expression Analysis (41)
- Homo sapiens (30)
- Mass spectrometry data (25)
- Multi-Omics (25)
- MERS-CoV (16)
- Mus musculus (15)
- Viruses (15)
- Health (14)
- Virus (14)
- Soil Microbiology (13)
- West Nile virus (11)
- Synthetic (10)
- Ebola (9)
- sequencing (9)
- Resource Metadata (8)
- Mass Spectrometry (7)
- Metagenomics (7)
- Microarray (5)
- Omics (5)
- PerCon SFA (5)
- Human Interferon (4)
- Microbiome (4)
- Proteomics (4)
- soil microbiology (3)
PerCon SFA, Co-Investigator Vivian Lin earned her PhD in organic chemistry from the University of California, Berkeley with Professor Chris Chang, developing fluorescent probes for imaging redox active small molecules. Afterward, she traveled to Switzerland for a postdoctoral fellowship in the...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM103 The purpose of this experiment was to evaluate the host response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004 (VN1203) virus, mutant VN1203-NS1trunc124, and mock...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL103 The purpose of this experiment was to evaluate the human host response to Influenza A virus (subtype H5N1) and mutant viruses. Sample data was obtained for human lung adenocarcinoma cell line (Calu-3)...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL104 The purpose of this experiment was to evaluate the host response to pandemic Influenza A virus (subtype H1N1) , natural isolate Influenza A/California/04/2009 virus. Sample data was obtained from human lung...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL102 The purpose of this experiment was to evaluate the human host cellular response to Influenza A virus (subtype H7N9) , wild-type strain Influenza A/Anhui/1/2013 (AH1-WT), and mutant viruses NS1-L103F/I106M...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM101 The purpose of this experiment was to evaluate the host mouse response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004, Influenza A/Vietnam/1203/2004 mutant strains PB2-627E...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM102 The purpose of this experiment was to evaluate the mouse host response to Influenza A virus (subtype H7N9) wild-type strain Influenza A/Anhui/1/2013 (AH1-WT) virus and mutants NS1-103F/106M (AH1-F/M) and...
Category
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL105 Metadata The purpose of this experiment was to evaluate the host epigenetic response to Influenza A virus (subtype H5N1) infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL106 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to Influenza A virus (subtype H5N1) infection. Samples were obtained from human lung adenocarcinoma cell line (Calu...
Category
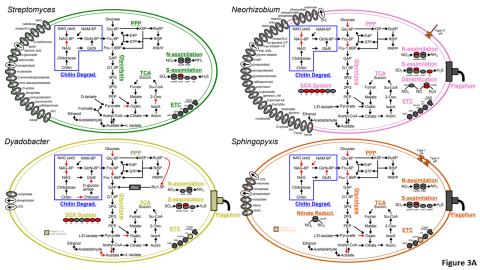
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...