Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MHAE003 The purpose of this experiment was to evaluate the human host response to wild-type MERS-CoV (strain EMC-2012) infection. Sample data was obtained from primary human airway epithelial cells (HAE) for mRNA...
Filter results
Tags
- (-) MERS-CoV (18)
- (-) Mass Spectrometry (14)
- (-) Synthetic (14)
- (-) PerCon SFA (10)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Viruses (26)
- Omics (25)
- Health (23)
- Virus (23)
- Soil Microbiology (21)
- Mus musculus (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- Ebola (11)
- Influenza A (11)
- High Throughput Sequencing (9)
- Metagenomics (9)
- Resource Metadata (9)
- Microbiome (8)
- Proteomics (8)
- Machine Learning (7)
- Microarray (7)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MM001 The purpose of this experiment was to evaluate the host response to wild-type MERS-CoV virus infection. Sample data was obtained from primary mouse (strain C57BL/6J) whole lung for mRNA, proteomics, metabolomics, and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MHAE002 The purpose of this experiment was to evaluate the human host response to wild-type MERS-CoV (strain EMC-2012) virus infection. Sample data was obtained from primary human microvascular endothelial cells (HMVE) for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL003 The purpose of this experiment was to evaluate the human host response to wild-type infectious clone of Middle Eastern Respiratory Syndrome coronavirus (icMERS-CoV) infection and mockulum. Sample data was obtained...
Category
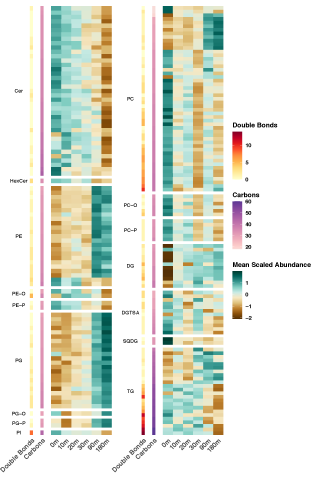
Rapid remodeling of the soil lipidome in response to a drying-rewetting event - Multi-Omics Data Package DOI Data package contents reported here are the first version and contain pre- and post-processed data acquisition and subsequent downstream analysis files using various data source instrument...
Category
Last updated on 2023-05-02T18:08:23+00:00 by LN Anderson Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68" Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
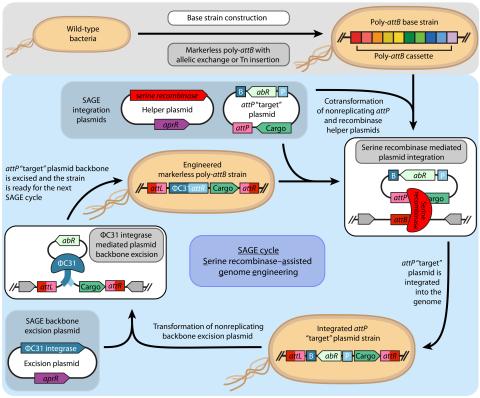
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
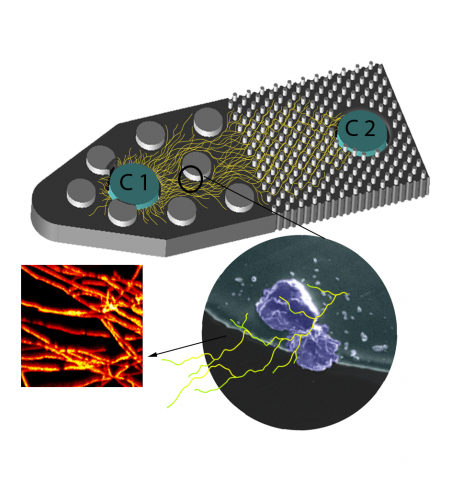
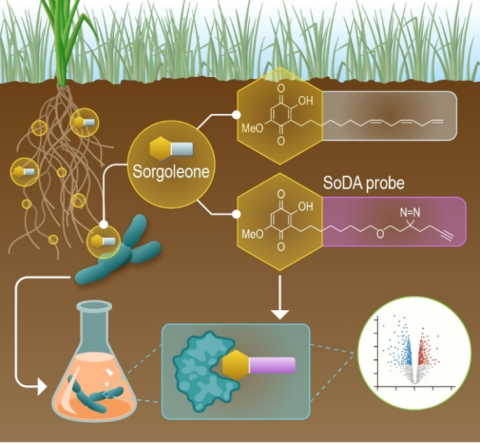
Last updated on 2024-04-19T19:12:08+00:00 by LN Anderson PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...