Comprised of 6,426 sample runs, The Environmental Determinants of Diabetes in the Young (TEDDY) proteomics validation study constitutes one of the largest targeted proteomics studies in the literature to date. Making quality control (QC) and donor sample data available to researchers aligns with...
Filter results
Category
- (-) Scientific Discovery (404)
- Biology (287)
- Earth System Science (167)
- Human Health (113)
- Integrative Omics (96)
- Microbiome Science (50)
- National Security (32)
- Computational Research (25)
- Computing & Analytics (18)
- Energy Resiliency (14)
- Chemical & Biological Signatures Science (12)
- Weapons of Mass Effect (12)
- Materials Science (11)
- Chemistry (10)
- Data Analytics & Machine Learning (9)
- Renewable Energy (8)
- Computational Mathematics & Statistics (7)
- Data Analytics & Machine Learning (7)
- Atmospheric Science (6)
- Ecosystem Science (6)
- Visual Analytics (6)
- Coastal Science (4)
- Energy Storage (4)
- Plant Science (4)
- Solar Energy (4)
- Bioenergy Technologies (3)
- Energy Efficiency (3)
- Transportation (3)
- Cybersecurity (2)
- Distribution (2)
- Electric Grid Modernization (2)
- Grid Cybersecurity (2)
- Subsurface Science (2)
- Water Power (2)
- Wind Energy (2)
- Advanced Lighting (1)
- Computational Mathematics & Statistics (1)
- Environmental Management (1)
- Federal Buildings (1)
- Geothermal Energy (1)
- Grid Analytics (1)
- Grid Energy Storage (1)
- High-Performance Computing (1)
- Terrestrial Aquatics (1)
- Vehicle Technologies (1)
- Waste Processing (1)
Tags
- Virology (77)
- Immune Response (51)
- Time Sampled Measurement Datasets (51)
- Differential Expression Analysis (46)
- Gene expression profile data (45)
- Homo sapiens (42)
- Predictive Phenomics (36)
- Multi-Omics (33)
- Mass spectrometry data (32)
- Viruses (27)
- Omics (26)
- Mass Spectrometry (25)
- Soil Microbiology (24)
- Health (23)
- Virus (23)
- MERS-CoV (19)
- Mus musculus (19)
- Proteomics (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- High Throughput Sequencing (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- S. elongatus PCC 7942 (10)
- Ebola (9)
- Microbiome (8)
- Post-Translational Modifications (8)
- Microarray (7)
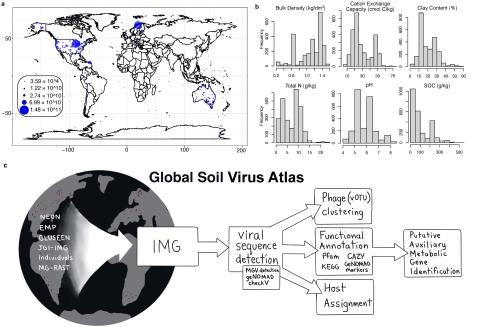
Please cite as : Graham E.B., A. Camargo, R. Wu, R.Y. Neches, M. Nolan, W.C. Nelson, and M.M. Pleake, et al. 2024. Global Soil Virus (GSV) Atlas. [Data Set] PNNL DataHub. 10.25584/2229733 This data is published under a CC0 license . The authors encourage data reuse and request attribution by...
Category
Please cite as : Wu R., A.E. Zimmerman, and K.S. Hofmockel. 2023. RNA viruses in grassland soils (Prosser, WA). [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33706 The data is comprised of RNA viral sequences that were bioinformatically screened from the de novo assemblies of...
Category
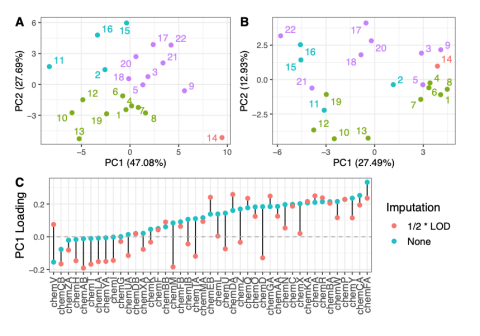
This data is supplementary to the manuscript Expanding the access of wearable silicone wristbands in community-engaged research through best practices in data analysis and integration by Lisa M. Bramer, Holly M. Dixon, David J. Degnan, Diana Rohlman, Julie B. Herbstman, Kim A. Anderson, and Katrina...
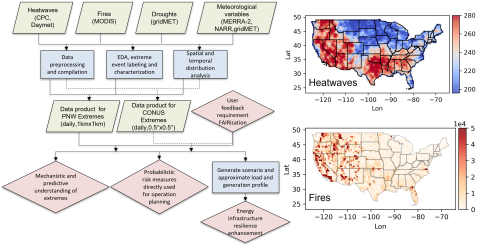
Extreme weather events, including fires, heatwaves(HWs), and droughts, have significant impacts on earth, environmental, and power energy systems. Mechanistic and predictive understanding, as well as probabilistic risk assessment of these extreme weather events, are crucial for detecting, planning...
A total of 172 children from the DAISY study with multiple plasma samples collected over time, with up to 23 years of follow-up, were characterized via proteomics analysis. Of the children there were 40 controls and 132 cases. All 132 cases had measurements across time relative to IA. Sampling was...
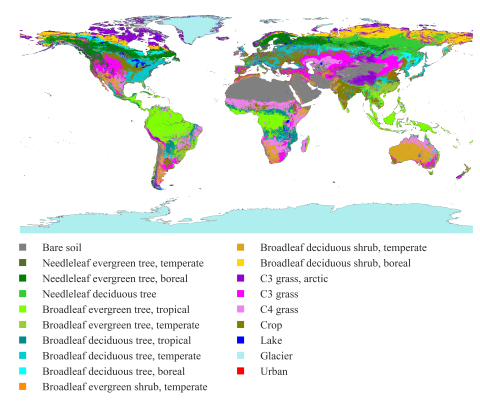
This dataset presents land surface parameters designed explicitly for global kilometer-scale Earth system modeling and has significant implications for enhancing our understanding of water, carbon, and energy cycles in the context of global change. Specifically, it includes four categories of...
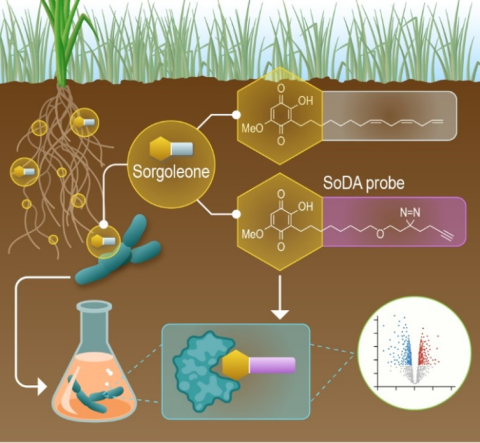
Created on 2024-09-17 by LN Anderson ; Last updated 2025-12-01 PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...
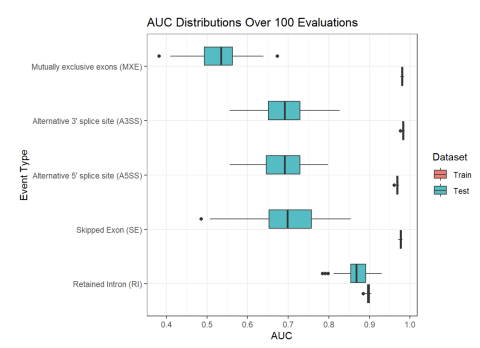
Inclusion levels of alternative splicing (AS) events of five different varieties (i.e. skipped exon (SE), retained intron (RI), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), and mutually exclusive exons (MXE)) were measured in human blood samples from two separate cohorts of...
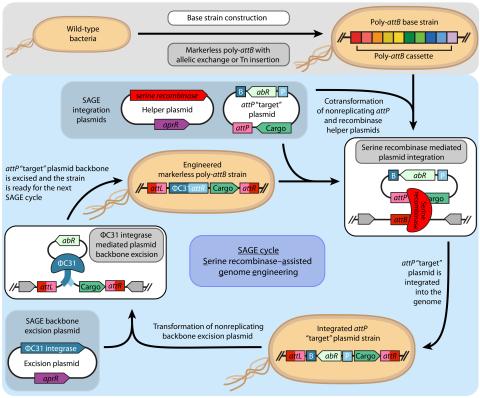
Last updated on 2023-03-21T18:35:22+00:00 by LN Anderson SAGE-RTP RT-PCR Amplicon Sequencing Barcode Count Analysis Promoter expression data for five bacterial species associated with the Serine recombinase Assisted Genome Engineering (SAGE) research project. Raw Measurement Data NCBI BioProject...
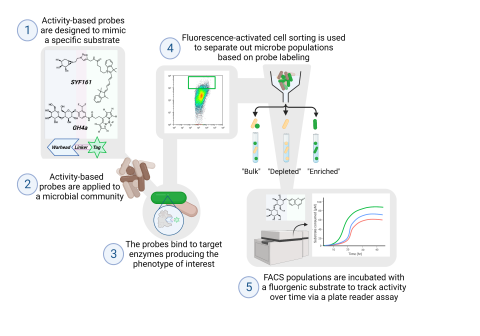
Last updated on 2025-08-11 by LN Anderson Activity-Based Probe Fluorescence-Activated Cell Sorting (ABP-FACS) of Soil Microbes (NR-DP0) Soil samples were incubated with either 0.2% cellobiose for three days or 0.2% xylan for nine days. Microbes were extracted from the soil matrix, incubated...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
Omics-LHV, West Nile Experiment WCD003 The purpose of this West Nile experiment was to obtain samples for omics analysis in mouse dendritic cell response to wild-type West Nile virus (WNV). Overall Design: Mouse dendritic cells (2 x 10^5) were treated with wild-type WNV and collected in parallel...