Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MHAE002 The purpose of this experiment was to evaluate the human host response to wild-type MERS-CoV (strain EMC-2012) virus infection. Sample data was obtained from primary human microvascular endothelial cells (HMVE) for...
Filter results
Content type
Tags
- (-) MERS-CoV (18)
- (-) Genomics (12)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Viruses (26)
- Omics (25)
- Health (23)
- Virus (23)
- Soil Microbiology (21)
- Mus musculus (18)
- Mass Spectrometry (14)
- Synthetic (14)
- sequencing (13)
- West Nile virus (13)
- Ebola (11)
- Influenza A (11)
- PerCon SFA (10)
- High Throughput Sequencing (9)
- Metagenomics (9)
- Resource Metadata (9)
- Microbiome (8)
- Proteomics (8)
- Machine Learning (7)
- Microarray (7)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL001 The purpose of this experiment was to evaluate the human host response to wild-type Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV), and mutants icMERS-CoV-RFP, icMERS-CoV-dNSP16, icMERS-CoV-d4B, and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL002 The purpose of this experiment was to evaluate the human host response to wild-type Middle Eastern Respiratory Syndrome coronavirus (MERS-CoV) and mutants icMERS-RFP, icMERS-DNSP16, and icMERS-d4B virus infection...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MFB001 The purpose of this experiment was to evaluate the human host response to wild-type MERS-CoV (icMERS-CoV) virus infection. Sample data was obtained from primary human fibroblasts and processed for mRNA, miRNA...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MMVE002 The purpose of this experiment was to evaluate the human host response to MERS-CoV (strain EMC-2012) infectious clone (icMERS-CoV) virus infection. Sample data was obtained from primary human fibroblast cells for...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MMVE002 The purpose of this experiment was to evaluate the human host response to MERS-CoV (strain EMC-2012) infectious clone (icMERS-CoV) virus infection. Sample data was obtained from primary human microvascular...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MM001 The purpose of this experiment was to evaluate the host response to wild-type MERS-CoV virus infection. Sample data was obtained from primary mouse (strain C57BL/6J) whole lung for mRNA, proteomics, metabolomics, and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MMVE001 The purpose of this experiment was to evaluate the human host response to MERS-CoV (strain EMC-2012) infectious clone (icMERS-CoV) virus infection. Sample data was obtained from primary human microvascular...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MHAE003 The purpose of this experiment was to evaluate the human host response to wild-type MERS-CoV (strain EMC-2012) infection. Sample data was obtained from primary human airway epithelial cells (HAE) for mRNA...
Category
Category
Omics Lethal Human Virus, SARS-CoV Experiment SM001 New uploads pending The purpose of this experiment was to evaluate the human host response to Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) wild-type virus. Sample data was obtained for 20 week-old C57BL/6J mouse lung tissue infected...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL003 The purpose of this experiment was to evaluate the human host response to wild-type infectious clone of Middle Eastern Respiratory Syndrome coronavirus (icMERS-CoV) infection and mockulum. Sample data was obtained...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
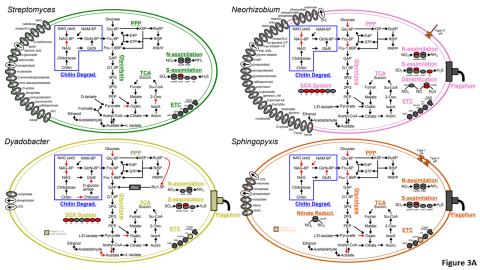
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
Last updated on 2023-05-31T16:35:53+00:00 by LN Anderson PerCon SFA: Sequencing of Sorgoleone Promoting Rhizobacteria Isolates Whole genome sequencing (WGS) of sorgoleone utilizing rhizobacteria strains Pseudomonas sorgoleonovorans SO81 , Burkholderia anthina SO82 , and Acinetobacter pittii SO1 , as...