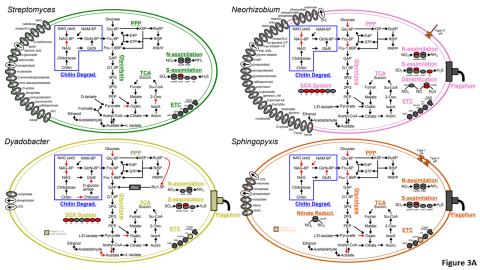
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
23