Biography Dr. Smith's research interests span the development of advanced analytical methods and instrumentation, with particular emphasis on high-resolution separations and mass spectrometry, and their applications in biological and biomedical research. This has included creating and applying new...
Filter results
Tags
- (-) Viruses (26)
- (-) Soil Microbiology (21)
- (-) Mass Spectrometry (14)
- (-) Metagenomics (9)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Omics (25)
- Health (23)
- Virus (23)
- MERS-CoV (18)
- Mus musculus (18)
- Synthetic (14)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- Ebola (11)
- Influenza A (11)
- PerCon SFA (10)
- High Throughput Sequencing (9)
- Resource Metadata (9)
- Microbiome (8)
- Proteomics (8)
- Machine Learning (7)
- Microarray (7)
Biography Kristin Burnum-Johnson is a senior scientist and team lead of the Biomolecular Pathways team at PNNL. Burnum-Johnson earned her PhD in Biochemistry from Vanderbilt University with Professor Richard M. Caprioli and then completed a postdoctoral fellowship at PNNL with Dr. Richard D. Smith...
Category
Biography Ernesto Nakayasu is a senior research scientist focused on understanding molecular mechanisms of diseases. Nakayasu has been applying systems biology and mass spectrometry-based omics measurements to study how pathogens and metabolic alterations cause human diseases; the goal is to...
Category
The IONTOF TOF.SIMS 5 data source is a time-of-flight secondary ion mass spectrometer and powerful surface analysis tool used to investigate scientific questions in biological, environmental, and energy research. Among the most sensitive of surface analysis tools, it uses a high-vacuum technique...
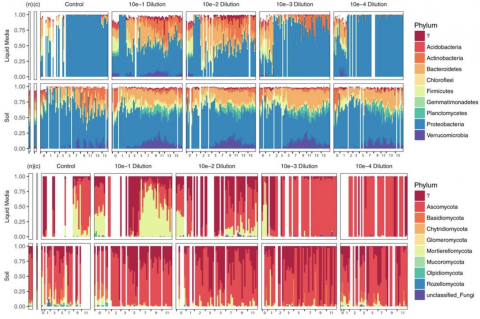
Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Category
Category
Category
Category
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL005 New uploads pending The purpose of this experiment was to evaluate the human host response to wild type virus icSARS-CoV Urbani and mutant virus icSARS deltaORF6 (DORF6) infection for subsequent transcriptional and proteomic analysis in...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL006 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome and proteome analysis in human lung 2B4 cells (clonal derivative of Calu-3 cells) infected with wild type icSARS CoV Urbani and icSARS Bat...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SCL008 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome and proteome analysis in human lung 2B4 cells (clonal derivative of Calu-3 cells) infected with either icSARS CoV, icSARS-deltaNSP16 or...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SM003 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome and proteome analysis in C57BlL/6 mice lung infected with dicSARS CoV, SARS MA15 wild type and SARS BatSRBD mutant virus. Overall Design...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SM012 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome and proteome analysis in C57Bl/6 mice infected with SARS MA15 or SARS deltaORF6 mutant viruses. Overall Design: Twenty-week-old C57BL/6 mice...
Category
Systems Virology Lethal Human Virus, SARS-CoV Experiment SHAE004 New uploads pending The purpose of this SARS experiment was to obtain samples for transcriptome analysis in Human Airway Epithelial (HAE) cells infected with SARS-CoV, SARS deltaORF6, SARS BatSRBD mutants in a longitudinal study...