Please cite as : Anderson L.N., W.C. Nelson, J.E. McDermott, R. Wu, S.J. Fansler, Y. Farris, and J.K. Jansson, et al. 2020. WA-TmG.1.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG1/1635002 To enable a comprehensive survey of the metabolic potential of complex soil...
Filter results
Category
Content type
Tags
- (-) Soil Microbiology (21)
- (-) MERS-CoV (18)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Viruses (26)
- Omics (25)
- Health (23)
- Virus (23)
- Mus musculus (18)
- Mass Spectrometry (14)
- Synthetic (14)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- Ebola (11)
- Influenza A (11)
- PerCon SFA (10)
- High Throughput Sequencing (9)
- Metagenomics (9)
- Resource Metadata (9)
- Microbiome (8)
- Proteomics (8)
- Machine Learning (7)
- Microarray (7)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL003 The purpose of this experiment was to evaluate the human host response to wild-type infectious clone of Middle Eastern Respiratory Syndrome coronavirus (icMERS-CoV) infection and mockulum. Sample data was obtained...
Category
Category
Last updated on 2023-05-02T18:08:23+00:00 by LN Anderson Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68" Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. Iso-VIG14.1.0 (Metagenome Derived Viral Genomes, WA/IA/KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/IsoVIG14/1770369 Soil samples were collected in triplicate in the Fall of 2017...
Category
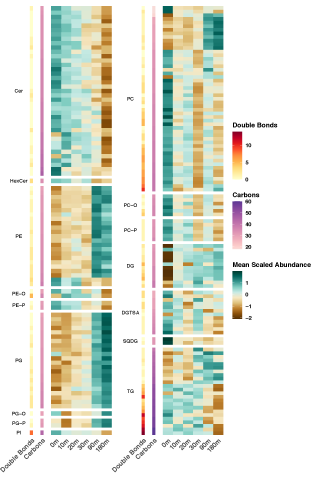
Rapid remodeling of the soil lipidome in response to a drying-rewetting event - Multi-Omics Data Package DOI Data package contents reported here are the first version and contain pre- and post-processed data acquisition and subsequent downstream analysis files using various data source instrument...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. IA-TmG.2.0 (Metagenome, IA). [Data Set] PNNL DataHub. https://doi.org/10.25584/IATmG2/1770333 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
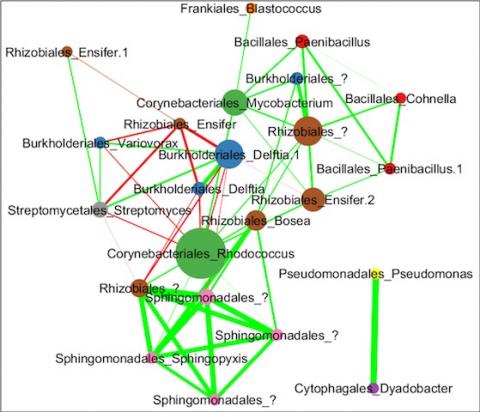
Please cite as : Anderson L.N., J.E. McDermott, and R.S. McClure. 2020. WA-IsoC_MSC1.1.0 (Amplicon 16S rRNA, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WAIsoCMSC1/1635272 The soil microbiome is central to the cycling of carbon and other nutrients and to the promotion of plant growth...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. WA-TmG.2.0 (Metagenome, WA). [Data Set] PNNL DataHub. https://doi.org/10.25584/WATmG2/1770324 Soil samples were collected in triplicate in the fall of 2017 across the three grassland...
Category
Please cite as : Anderson L.N., R. Wu, W.C. Nelson, J.E. McDermott, K.S. Hofmockel, and J.K. Jansson. 2021. KS-TmG.2.0 (Metagenome, KS). [Data Set] PNNL DataHub. https://doi.org/10.25584/KSTmG2/1770332 Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations...
Category
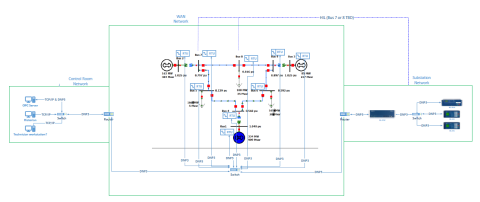
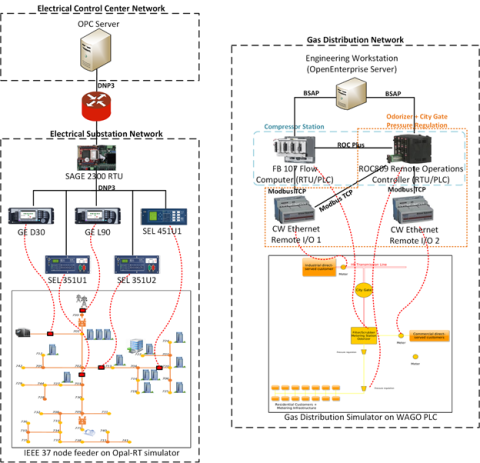
This dataset includes one baseline and three cybersecurity based scenarios utilizing the IEEE 9 Bus Model. This instantiation of the IEEE 9 model was built utilizing the OpalRT Simulator ePhasorsim module, with Bus 7 represented by hardware in the loop (HiL). The HiL was represented by two SEL351s...
This dataset includes the results of high-fidelity, hardware in the loop experimentation on simulated models of representative electric and natural gas distribution systems with real cyber attack test cases. Such datasets are extremely important not only in understanding the system behavior during...
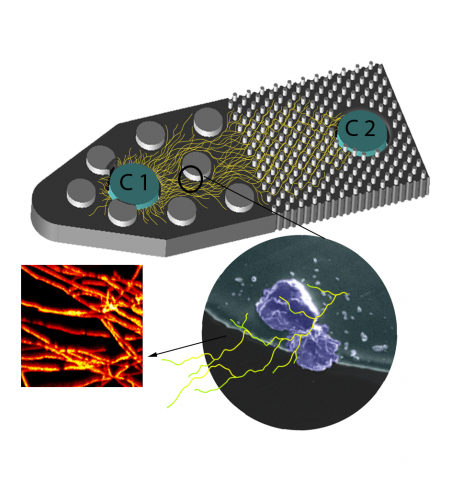
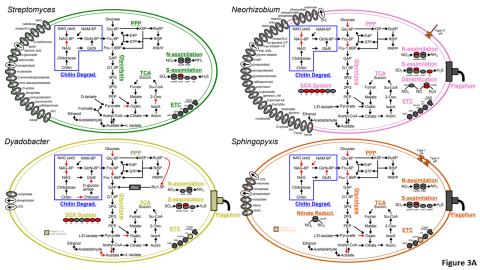
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes. [Data Set] PNNL DataHub. https://doi.org/10.25584/PNNLDH/1986536 Model Soil Consortium 2 (MSC-2) Bacterial Isolate Genomes...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL004 Metadata The purpose of this experiment was to evaluate the host response to MERS-CoV virus infection. Sample data was obtained from human lung adenocarcinoma cells (Calu-3) and processed for genome binding/occupancy...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL005 Metadata The purpose of this experiment was to evaluate the human host epigenetic response to MERS-CoV virus infection. Samples were obtained from human lung adenocarcinoma cells (Calu-3) and processed for...