Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM101 The purpose of this experiment was to evaluate the host mouse response to Influenza A virus (subtype H5N1) wild-type strain Influenza A/Vietnam/1203/2004, Influenza A/Vietnam/1203/2004 mutant strains PB2-627E...
Filter results
Content type
Tags
- (-) West Nile virus (13)
- (-) Proteomics (8)
- (-) Microarray (7)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Multi-Omics (30)
- Viruses (27)
- Omics (26)
- Health (23)
- Virus (23)
- Soil Microbiology (22)
- MERS-CoV (18)
- Mus musculus (18)
- Mass Spectrometry (14)
- Synthetic (14)
- sequencing (13)
- Genomics (12)
- Ebola (11)
- Influenza A (11)
- Metagenomics (10)
- PerCon SFA (10)
- High Throughput Sequencing (9)
- Resource Metadata (9)
- Microbiome (8)
- Machine Learning (7)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCT001 The purpose of this experiment was to evaluate the host responseto West Nile virus (WNV-NY99) wild-type (strain 382) and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WDC010 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from primary mouse...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WDC011 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from primary mouse...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WGCN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WGCN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WSE001 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 virus infection. Sample data was obtained from mouse (strain C57BL6/JAX) blood serum...
Category
Biomedical Resilience & Readiness in Adverse Operating Environments (BRAVE) Project: Exhaled Breath Condensate (EBC) TMT Proteomic Transformation Data Exhaled breath condensate (EBC) represents a low-cost and non-invasive means of examining respiratory health. EBC has been used to discover and...
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WLN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WLN002 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
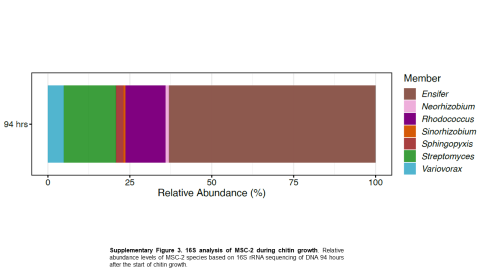
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. 16s data from MSC-2 growth. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33231 16s data from MSC-2 growth 3 fastq of 16s amplicon data of MSC2 1 csv file of raw...
Category
Please cite as : McClure R.S., Y. Farris, R.E. Danczak, W.C. Nelson, H. Song, A. Kessler, and J. Lee, et al. 2022. Metatranscriptomic data from MSC-2. [Data Set] PNNL DataHub. https://data.pnnl.gov/group/nodes/dataset/33232 Metatranscriptomic data from MSC-2 12 fastq files (6 forward read, 6 reverse...
Category
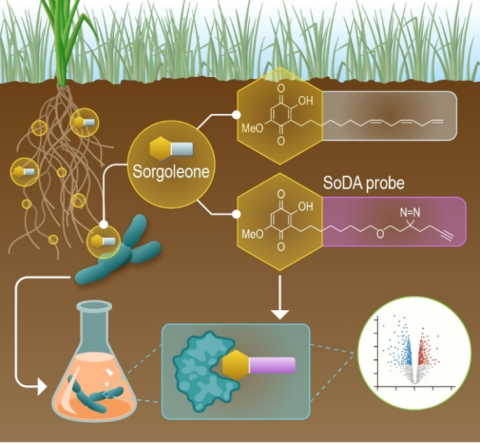
Last updated on 2024-04-19T19:12:08+00:00 by LN Anderson PerCon SFA: Profiling sorghum-microbe interactions with a specialized photoaffinity probe identifies key sorgoleone binders in Acinetobacter pitti Mass spectrometry-based global proteome analysis and SoDA-PAL photoaffinity probe labeled...