Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL104 The purpose of this experiment was to evaluate the host response to pandemic Influenza A virus (subtype H1N1) , natural isolate Influenza A/California/04/2009 virus. Sample data was obtained from human lung...
Filter results
Content type
Tags
- (-) Multi-Omics (30)
- (-) Mass Spectrometry (13)
- Virology (79)
- Immune Response (53)
- Time Sampled Measurement Datasets (50)
- Gene expression profile data (47)
- Differential Expression Analysis (46)
- Homo sapiens (34)
- Mass spectrometry data (31)
- Viruses (26)
- Omics (24)
- Health (23)
- Virus (23)
- Soil Microbiology (21)
- MERS-CoV (18)
- Mus musculus (18)
- sequencing (13)
- West Nile virus (13)
- Genomics (12)
- Ebola (11)
- Influenza A (11)
- PerCon SFA (10)
- High Throughput Sequencing (9)
- Metagenomics (9)
- Resource Metadata (9)
- Microbiome (8)
- Proteomics (8)
- Machine Learning (7)
- Microarray (7)
- Synthetic Biology (7)
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment ICL102 The purpose of this experiment was to evaluate the human host cellular response to Influenza A virus (subtype H7N9) , wild-type strain Influenza A/Anhui/1/2013 (AH1-WT), and mutant viruses NS1-L103F/I106M...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Influenza A Virus Experiment IM102 The purpose of this experiment was to evaluate the mouse host response to Influenza A virus (subtype H7N9) wild-type strain Influenza A/Anhui/1/2013 (AH1-WT) virus and mutants NS1-103F/106M (AH1-F/M) and...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WCT001 The purpose of this experiment was to evaluate the host responseto West Nile virus (WNV-NY99) wild-type (strain 382) and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain C57BL...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Ebola Virus Experiment EHUH002 The purpose of this experiment was to evaluate the human host response to wild-type Zaire Ebola virus (strain Mayinga) and mutant virus infection. Samples were obtained from human hepatoma carcinoma cells (HUH-7)...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson MERS-CoV Experiment MCL003 The purpose of this experiment was to evaluate the human host response to wild-type infectious clone of Middle Eastern Respiratory Syndrome coronavirus (icMERS-CoV) infection and mockulum. Sample data was obtained...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson Ebola Virus Experiment EH001 The purpose of this experiment was to evaluate the human patient peripheral blood mononuclear cells (PBMC) response to Zaire Ebola virus (strain Makona) infection during the 2013-2016 epidemic in West Africa...
Category
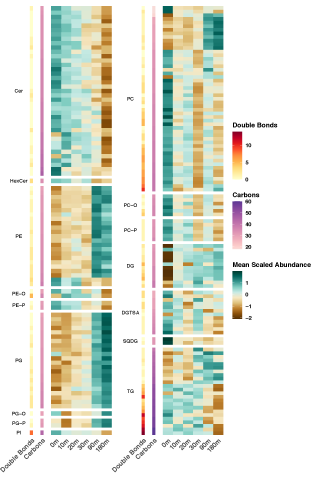
Rapid remodeling of the soil lipidome in response to a drying-rewetting event - Multi-Omics Data Package DOI Data package contents reported here are the first version and contain pre- and post-processed data acquisition and subsequent downstream analysis files using various data source instrument...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WLN003 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A 2 nt virus infection. Sample data was obtained from mouse (strain...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WDC010 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 and mutant 382-E218A virus infection. Sample data was obtained from primary mouse...
Category
Last updated on 2024-02-11T22:41:43+00:00 by LN Anderson West Nile Virus Experiment WSE001 The purpose of this experiment was to evaluate the host response to West Nile virus (strain WNV-NY99) wild-type clone 382 virus infection. Sample data was obtained from mouse (strain C57BL6/JAX) blood serum...
Category
Last updated on 2023-05-02T18:08:23+00:00 by LN Anderson Fungal Monoisolate Multi-Omics Data Package DOI "KS4A-Omics1.0_FspDS68" Molecular mechanisms underlying fungal mineral weathering and nutrient translocation in low nutrient environments remain poorly resolved, due to the lack of a platform for...
Category
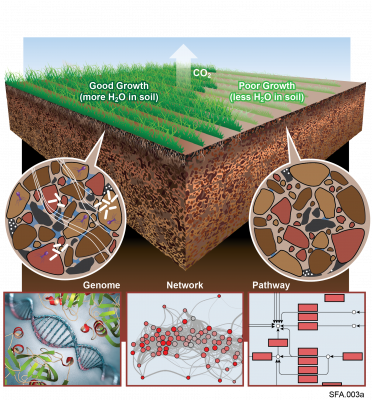
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome...
Datasets
23
LIQUID Software Overview LIQUID provides users with the capability to process high throughput data and contains a customizable target library and scoring model per project needs. The graphical user interface provides visualization of multiple lines of spectral evidence for each lipid identification...
Category
pmartR Software Overview The pmartR package provides a single software tool for QC (filtering and normalization), exploratory data analysis (EDA), and statistical analysis (robust to missing data) and includes numerous visualization capabilities of mass spectrometry (MS) omics data (proteomic...