Description
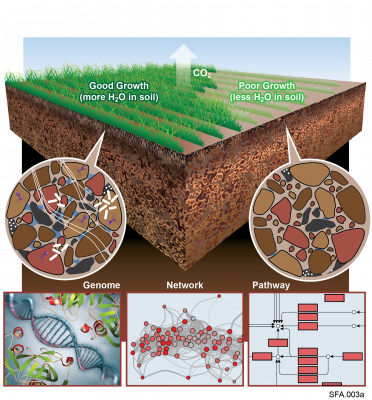
The Phenotypic Response of the Soil Microbiome to Environmental Perturbations Project (Soil Microbiome SFA) at Pacific Northwest National Laboratory is a Genomic Sciences Program Science Focus Area (SFA) Project operating under the Environmental Microbiome Science Research Area. The Soil Microbiome SFA aims to develop a systems-level understanding of soil microbial and phenotypic responses to changing moisture through a tractable, spatially explicit examination of the molecular and ecological interactions occurring within and between microbial consortia in soil. The Soil Microbiome SFA project repository at PNNL DataHub allows for exploring and downloading integrated experimental omics data, experimental metadata, pre- and post-processed data files, and other associated materials directly related to experimental project publication data.
For updates and more information, visit the PNNL Soil Microbiome SFA homepage.
Related Resources
- Program Homepage: Phenotypic Response of Soil Microbiomes
- KBase Collaboration: Metabolic Network Reconstruction of Microorganisms and Communities
Federal Acknowledgements
This research was supported by the Department of Energy Office of Biological and Environmental Research (BER) and is a contribution of the Scientific Focus Area ‘Phenotypic response of the soil microbiome to environmental perturbations’. PNNL is operated for the DOE by Battelle Memorial Institute under Contract DE-AC05-76RLO1830.
(This page was prepared by LN Anderson)
Last updated on 2024-06-03